Abstract
Purpose
Development of a polyethylene glycol (PEG)-stabilized immunoliposome (PSIL) formulation with high DNA content suitable for in vivo intravenous administration and targeted gene delivery.
Materials and Methods
Plasmid DNA was condensed using 40% ethanol and packaged into neutral PSILs targeted to the mouse transferrin receptor using monoclonal antibodies (MAbs; clones RI7 and 8D3) attached to their PEG maleimide moieties. PSILs size was measured by quasi-elastic light scattering. The targeting capacity of the formulation was determined by transfection of mouse neuroblastoma Neuro 2A (N2A) cells with PSIL-DNA complexes conjugated with either RI7 or 8D3 MAbs.
Results
DNA encapsulation and MAb conjugation efficiencies averaged 71 ± 14% and 69 ± 5% (mean ± SD), respectively. No alteration in mean particle size (< 100 nm) or DNA leakage were found after 48 h storage in a physiological buffer, and the in vivo terminal half-life reached 23.9 h, indicating that the PSIL-DNA formulation was stable. Addition of free RI7 MAbs prevented transfection of N2A cells with PSIL-DNA complexes conjugated with either RI7 or 8D3 MAbs, confirming that the transfection was transferrin receptor-dependent.
Conclusions
The present data suggest that our new PSIL formulation combines molecular features required for targeted gene therapy including high DNA encapsulation efficiencies and vector-specific transient transfection capacity.
Similar content being viewed by others
INTRODUCTION
Gene therapy depends on efficient and selective vector systems to deliver gene medicine into target cells. Owing to effective transfection and long-lasting expression of exogenous genes into cells, viral vectors became the method of choice for brain gene therapy in academic research laboratories (1–5). However, most of the available viral vectors are associated with major safety problems such as mutagenesis due to long-term integration into the genome (retrovirus or adeno-associated virus, AAV) or serious inflammatory responses (herpes simplex virus-1, HSV-1, or adenovirus, AV), making the translation into clinical applications hazardous (4–10).
Liposome-based non-viral DNA formulations have been recently developed and might offer a safer alternative to viral vectors (10–13). Indeed, expression plasmids can be complexed within liposomes to transfect cells in vitro (14–16) and in vivo (17–20). Liposomes are composed of naturally-occurring, biodegradable lipids organized into bilayer membranes surrounding an aqueous core, and thus display a low level of immunogenicity or toxicity (11,12). In contrast with viral vector-mediated transfection, expression plasmids transfected using liposomes do not normally integrate into the genome, but rather undergo episomal expression as non-replicating extrachromosomal elements in the nucleus (15,17). Therefore, transfection using liposome-DNA complexes is expected to be reversible and to allow repeated administrations, which is a major advantage from a clinical standpoint. Further data indicate that selection of promoters controlling the therapeutic gene can restrict its expression to a specific type of cell (18).
To be suitable for in vivo gene therapy, a candidate liposome formulation must combine cell transfection and drug delivery capacity, and thus requires many important features. First, adding polyethylene glycol (PEG) tails to liposomes strikingly decreases the systemic plasma clearance, enhancing the apparent terminal half-life of PEG-stabilized liposomes (PSLs) to up to 90 h in humans (21–24). Liposome nanoparticles also offer a haven for encapsulated DNA after systemic injections, and PSL-DNA complexes show a much better pharmacokinetic profile than naked DNA (11,12,25). However, the size of PEG strands must be kept under 2,000 Da to allow binding of the conjugated proteic vector to its target (23,25). Several PSL formulations (“stealth” liposomes) are now approved for clinical use (12,25,26). Second, selecting a composition in phospholipids to obtain a neutral total charge within the liposome membranes prevents non-specific interactions with blood components and reduces entrapment in the lung and the liver, which is the usual fate of cationic liposomes (12,21,23,25,27–30). Third, the liposome formulation should allow a high conjugation efficiency to proteic vectors to form PEG-stabilized immunoliposomes (PSILs) suitable for selective drug delivery and, more specifically, for targeting of receptor-mediated transport/endocytosis systems in the organism (11,12,15,25,31). Fourth, the mean size of the particles must be under 100 nm to optimize their bioavailability in vivo (12,23,25). Besides not fulfilling all these requirements, most published liposome formulations show relatively low DNA encapsulation efficiencies, which consequently limit their transfection capacity (17,25,32–34). The objectives of the present work were thus to develop and characterize a new PSIL formulation with a high DNA content, and to test its gene delivery capacity in vitro.
MATERIALS AND METHODS
Materials
Unless otherwise noted, all reagents were obtained from Sigma-Aldrich (Oakville, ON, Canada). POPC (1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine) and DSPE (distearoylphosphatidylethanolamine)—PEG2000 (2,000 Da polyethylene glycol) were purchased from Northern Lipids Inc. (Vancouver, BC, Canada). DSPE-PEG2000-maleimide was synthesized by Nektar Therapeutics (Huntsville, AL). The hybridoma cell lines expressing rat monoclonal antibodies (MAbs) raised against the mouse transferrin receptor were gifts from Dr Jayne Lesley (Salk Institute, San Diego, CA) and Dr Pauline Johnson (University of British Columbia, Vancouver, BC, Canada) for clone RI7.217 and from Dr Britta Engelhardt (University of Bern, Switzerland) for clone 8D3. Competent cells were purchased from Stratagene (La Jolla, CA), [α-33P]-dCTP (3000 Ci/mmol) was from Perkin Elmer Life Sciences (Woodbridge, ON, Canada), N-succinimidyl-[2,3-3H] propionate (3H-NSP, 101 Ci/mmol), Sephadex G-25 and Sepharose CL-4B size exclusion gels were purchased from GE-Healthcare-Amersham (Baie d’Urfé, QC, Canada). Devices for MAb purification and sterile filtration were from Millipore (Cambridge, ON, Canada).
Production and Purification of Monoclonal Antibodies (MAbs) in Vitro
Hybridoma cell lines expressing rat MAbs raised against the mouse transferrin receptor (clones RI7.217 (RI7) and 8D3) were cultured in MAb serum-free medium in CELLine bioreactors (BD Biosciences, Mississauga, ON, Canada). Supernatants were harvested weekly. MAbs were purified using Montage Prosep-G spin columns according to the manufacturer’s recommendations. Purified antibodies were concentrated with Amicon (molecular weight cut-off, MWCO = 30 kDa) ultracentrifugal devices and subsequently dialyzed against 0.01 M PBS (pH 7.4) using Slide-A-Lyser dialysis cassettes (MWCO = 10 kDa, Pierce, Rockford, IL). Protein concentrations were determined using bicinchoninic acid assays (Pierce). Weekly yields of purified MAbs averaged 3 mg.
MAb Radiolabeling with [3H]-NSP
One mCi of [3H]-NSP was evaporated under a nitrogen stream in a borosilicate tube and incubated with 500 mg of MAbs into 100 μl of a buffer containing 0.1 M sodium borate / 1.5 M NaCl (pH 8.0) for 1 h at room temperature, followed by a 2-h incubation at 4°C. Radiolabeled MAbs were separated using a G-25 Sephadex gel filtration column pretreated with 10% (w/v) BSA, and specific activity was computed using disintegrations per minute (dpm) counts and protein content in eluted fractions.
Plasmids Production and Radiolabeling
The pGL3 (5.3 kb) and pGL4 (4.6 kb) plasmids expressing Renilla luciferase under the control of the simian virus 40 (SV40) promoter (Promega, Madison, WI), and the pCMV-GLuc (pGLuc) (5.8 kb) plasmid (New England Biolabs, Pickering, ON, Canada) driving the expression of Gaussia luciferase with the cytomegalovirus (CMV) promoter were compared. Plasmids were extracted after transformation using the GenEluteTM HP Endotoxin-Free Plasmid Maxiprep Kit (Sigma) and further purified by alcohol precipitation. A fraction of plasmids were then labeled with [33P]-dCTP using a nicktranslation system (Invitrogen, Burlington, ON, Canada). Radiolabeled plasmids were purified using a Strataprep PCR purification kit (Stratagene) and then used as tracers to monitor DNA encapsulation efficiencies into liposomes.
Preparation of the PSL Formulation and DNA Encapsulation
Briefly, POPC (18.6 μmol), DDAB (0.6 μmol), DSPE-PEG2000 (0.6 μmol), and the linker lipid DSPE-PEG2000-maleimide (0.2 μmol) were dissolved in chloroform, followed by evaporation under a nitrogen stream and constant agitation in order to produce a thin lipid film, which was allowed to dry during 30 min. This proportion of cationic (DDAB) and anionic (DSPE) lipids is expected to result in a net neutral charge at the surface of liposomes (33). To obtain non PEGylated liposomes, non PEGylated DSPE (0.8 μmol) was used instead in the same proportion. The dried lipid film was dispersed in 0.2 ml 0.05 M Tris-HCl (pH 7.0), vortexed for 2 min, followed by 2 min of bath sonication and a second 1-min vortex agitation. Unlabeled plasmids (100–250 μg) as well as ∼2 μCi of [33P]-labeled plasmid DNA were added to the lipids along with 0.1 M Tris-HCl buffer (pH 7.0) up to a volume of 0.4 ml. Then, 0.6 ml of a 67 % (v/v) ethanol solution in 0.05 M Tris-HCl buffer (pH 7.0) was added drop by drop during 30 sec under minimal agitation to reach a final volume of 1.0 ml. For comparison, series of liposomes were prepared in parallel experiments using freeze-thaw cycles as described previously (17,18). The resulting large vesicles were converted into liposomes under 100 nm by extrusion using a hand-held LiposoFast™-Basic extruder (Avestin, Ottawa, ON, Canada) as described (17,18). To remove unencapsulated plasmid DNA while encapsulated DNA remains protected by the liposomes, the PSL suspension was exposed to a 1-h nuclease digestion [6 units of bovine pancreatic deoxyribonuclease I and 33 units of exonuclease III in 5 mM MgCl2, 37°C) as previously shown (17,18,32).
MAb Conjugation to PSLs
Three mg of unlabeled MAbs (RI7 or 8D3) and 0.5 μCi of tritiated MAbs were thiolated with a 40: 1 molar excess of freshly prepared 2-iminothiolane (Traut’s reagent) following a 1-h incubation at room temperature in 0.05 M sodium borate/0.1 mM EDTA (pH 8.5). Thiolated MAbs were diluted 15 times in 0.05 M HEPES/0.1 mM EDTA (pH 7.0) and then concentrated using an Amicon filter device (MWCO = 30 kDa). This step was repeated twice to discard 2-iminothiolane, to transfer MAbs into HEPES buffer, and to reduce the final volume of the solution to less than 500 μl. To conjugate MAbs to PEG-maleimide strands on the PSLs, thiolated MAbs were added to the liposome-DNA preparation for an overnight room temperature incubation in a 2 ml glass bottle under inert nitrogen atmosphere.
Determination of DNA Encapsulation and MAb Conjugation Efficiencies using Size Exclusion Chromatography
Unconjugated MAbs and digested DNA were separated from the PSILs by size exclusion chromatography. After the overnight conjugation, PSILs were eluted with 0.05 M HEPES (pH 7.0) at a rate of 1.0 ml/min in a 1.5 cm × 20 cm pre-equilibrated Sepharose CL-4B column. Fractions were collected and quantified by liquid scintillation counting to determine their 3H and 33P radioisotopes content. DNA encapsulation efficiency was computed as the sum of 33P in the liposome peak divided by the sum of 33P in the digested DNA peak and the liposome peak (Fig. 1). Similarly, MAb conjugation efficiencies were computed as the sum of 3H in the liposome peak over the sum of 3H in free unconjugated MAb fractions and liposome fractions (Fig. 1). Conjugation of MAbs to the liposomes was also confirmed using bicinchoninic acid protein determination assays. All eluted fractions containing PSILs were kept in glass vials under nitrogen atmosphere and stored at 4°C unless specified otherwise.
Typical Sepharose CL-4B size exclusion chromatography profile showing that 76% of plasmid DNA was incorporated into the neutral unilamellar polyethylene glycol (PEG)-stabilized immunoliposomes and that 84% of RI7 monoclonal antibodies (MAbs) were attached to the liposomes. Trace amounts of radiolabeled [33P]-DNA and [3H]-RI7 MAb were included in the preparation to monitor encapsulation and conjugation efficiencies, respectively.
Electron Microscopy
Unconjugated PSL suspensions (8 μl) were applied to Formvar-coated 300 mesh copper grids (Canemco-Marivac, Montréal, QC, Canada), and allowed to settle for 3 min. Excess liquid was blotted off and the grids were washed twice with ultrapure water. The liposome preparation was then negatively stained with 1% (w/v) uranyl acetate (pH 4.5) for 2 min in triplicates. Grids were blotted off again and allowed to dry. Samples were examined using a JEM 1010 transmission electron microscope (JEOL, Peabody, MA) at a magnification of X 75,000 and X 100,000, and pictures were taken with a CCD camera (Gatan Inc, Warrendale, PA) and enlarged using Canvas X (ACD Systems, Saanichton, BC, Canada) on a G4 Power Macintosh.
Dynamic Light Scattering: Particle Size Measurement
Quasi-elastic light scattering (QELS) size determination of PSLs and PSILs was performed in duplicates using a Nicomp C-370 Particle Sizer (Pacific Scientific Hiac/Royco Instruments division, Particle Sizing Systems Inc., Santa Barbara, CA). Various dilutions of the samples in 0.05 M HEPES or Dulbecco’s Modified Eagle’s Medium (DMEM) (pH 7.0) were adjusted to reach an intensity set point of 300 kHz. Measurements were taken with the following fixed parameters : a temperature of 23°C, a liquid viscosity of 0.933 cP, a liquid index refraction of 1.333, a laser wavelength of 632.8 nm, a scattering angle of 90°, an initial delay time of 30 sec, and an analysis time of 5 min. Results were expressed as a Gaussian graph of the relative volume occupied by the particles in function of the size of liposomes, and the mean diameter was calculated. The distribution of the particles formed one single major population. Latex beads of 100 nm (Beckman-Coulter, Mississauga, ON, Canada) were used as standards to confirm the accuracy of the measurements.
Stability of the Liposome Preparations in Vitro and in Vivo
To determine the size stability of PSLs and PSILs, QELS measurements were performed as described above for formulations kept under a nitrogen atmosphere at room temperature or 4°C, in 0.05 M HEPES (pH 7.0), with or without 0.09% (w/v) NaCl. Measurements were taken in duplicates for each sample on day 1, 3, 6, 9 and 21 following liposome synthesis (Fig. 3a–b). In addition, to test the size stability of the particles in conditions closer those encountered in vivo, we performed QELS measurements of non PEGylated liposomes, PSLs and PSILs kept under a nitrogen atmosphere at room temperature or 37°C in DMEM for 24 or 48 h (Fig. 3c).
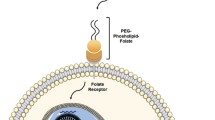
(a) Schematic representation of the polyethylene glycol (PEG)-stabilized immunoliposomes (PSILs) showing encapsulated DNA and conjugated monoclonal antibodies (MAbs). (b–d) Transmission electron microscopy view of the uranyl acetate stained PEG-stabilized liposomes (PSLs). Magnification bar = 100 nm. Size distributions of (e) PSLs and (f) PSILs according to quasi-elastic light scattering analyses.
To determine whether the DNA plasmids leaked from liposomes in physiological conditions, the PSIL formulation was kept in 0.05 M HEPES buffer or DMEM at 37°C. The percentage of DNA that remained entrapped in liposomes was computed after 24 and 48 h using size exclusion chromatography as described above. To estimate the terminal half-life of the [33P]-DNA encapsulated in PSILs, 2 μCi of the preparation were injected in the tail vein of three rats. Blood samples (∼180 μl) were drawn at 1.5, 15, 30 and 60 min and, then, at 8, 25, 46, and 96 h and centrifuged (10,000 g; 5 min) to collect plasma. Radioactivity counts were measured using liquid scintillation counting. The terminal half-life was determined from the slope of the linear part of the log(concentration) versus time curve using the formula t1/2 = log10(2)/slope (35).
DNA Analysis
In order to confirm the encapsulation efficiency and the integrity of plasmids encapsulated within the PSLs, the eluted fractions recovered after the size exclusion chromatography were concentrated with a Speed Vacuum machine (SpeedVac® Plus SC110 A, Savant Instruments Inc., Holbrook, NY), lysed at 50°C for 30 min with 1% (v/v) Triton X-100, 1% (w/v) sodium deoxycholate and 2% sodium dodecyl sulfate, and further concentrated with a DNA Clean and Concentrator™-5 kit (Zymo Research, Orange, CA) to discard detergent. Samples were then subjected to a 0.6% (w/v) agarose gel electrophoresis (migrated at 106 V for 1 h at room temperature) and observed with a UV transilluminator. To determine whether the ethanol needed for DNA encapsulation was interfering with nuclease digestion, pGL3 DNA plasmids (1 μg) were exposed to various amounts of ethanol (0–60%) and to nucleases as described above during 2 h at 37°C. Resulting samples were run on a 0.9% (w/v) agarose gel either right after the digestion or after being kept overnight at room temperature to mimic the conditions occurring during the conjugation procedures. Encapsulation efficiencies were computed using image analysis of UV densitometry (Alpha imager system, Alpha Innotech, San Leandro, CA).
In Vitro Transfection
To determine the relative capacity of two proteic vectors to carry the PSIL and its DNA content through the cell membrane, transfection experiments were performed with a cell line known to express the mouse transferrin receptor, the neuroblastoma Neuro 2A (N2A) mouse cell line (36,37). The presence of transferrin receptors on N2A cells was further confirmed with a Dot Blot analysis using the RI7 antibody (not shown). N2A cells were grown in serum-free VP-SFM medium with 4 mM L-glutamine supplemented with penicillin (10 U/ml) and streptomycin (10 μg/ml). The day before transfections, 2.0 × 104 cells per well were seeded in a 96-well microplate. Cells in each well were incubated with 160 ng of DNA encapsulated in unconjugated PSLs or PSILs conjugated with either RI7 or 8D3 MAbs, with or without free competing RI7 MAbs (final concentration: 0.5 mg/ml). After being concentrated using Amicon filter devices (MWCO = 100 kDa), the liposome preparations and free MAbs were filter-sterilized (0.22 μm) before use in cell culture. Cells were incubated at 37°C for 16 h in serum- and antibiotic-free culture medium and for a further 32 h in serum-free medium as described above. Cell lysates were then processed for luciferase activity measurements using the Steady-Glo Luciferase Assay reagent kit (Promega) for pGL3/4 plasmids, while supernatants were used with the Gaussia Luciferase Assay kit (New England BioLabs) for pGLuc, according to the manufacturer’s instructions. Luminescence data were generated using a 96-well microplate luminometer (Synergy™ HT, Biotek, Winooski, VT).
RESULTS
High Encapsulation Efficiencies of Plasmid DNA into PSILs
Owing to the capacity of ethanol to condense DNA (38,39), high encapsulation efficiencies (mean ± SD: 71 ± 14%) of plasmid DNA into PSILs were obtained, based on 58 experiments (Fig. 1 and Table I). Encapsulation efficiencies were similar between the commercial plasmids compared here (pGLuc, pGL3 and pGL4). The encapsulation efficiencies were superior to values obtained with a freeze-thaw method (Table I) and to most previously reported data (17,32,33,38,40). Also shown in Table I, the 40% ethanol-based methodology led to higher encapsulation efficiencies than lower ethanol percentages. In parallel experiments, it was found that varying the pH between 5 and 8, addition of 5% cholesterol in the lipid composition of the vesicles (with 88% POPC instead of the usual 93% POPC) or 1.5 mM calcium chloride during the plasmid encapsulation step did not significantly change the encapsulation efficiencies (data not shown).
High Conjugation Efficiencies of Proteic Vectors (transferrin receptor MAbs) to PSLs
Conjugation was performed by linking thiolated MAbs to the maleimide moieties of PEG2000 strands attached to the core of PSLs. The level of thiolation of MAbs can be controlled by varying the 2-iminothiolane : MAb ratio, and it was found that a ratio of 40: 1 led to conjugation efficiencies close to 70%, based on 23 experiments (Table I and Fig. 1). Assuming a MAb molecular weight of 150,000 kDa, a loss of 25% of lipids in the extrusion steps and 100,000 lipid molecules per liposome, it can be estimated that 92 MAbs were attached to any single liposome. A schematic representation of a PSIL is illustrated in Fig. 2a.
The mean diameter of the polyethylene glycol (PEG)-stabilized (a) liposomes (PSLs) and (b) immunoliposomes (PSILs) remained stable for up to 3 weeks after synthesis after storage at room temperature (RT) or at 4°C in 0.05 M HEPES buffer (pH 7.0), with or without 0.09% (w/v) NaCl. (c) Non PEGylated liposomes as well as PSLs) and PSILs remained stable for up to 48 h after synthesis when stored at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM). Mean particle sizes were measured over time using quasi-elastic light scattering analysis. Values are mean ± SD of at least three measurements.
Size and Stability of PSILs
The size and shape of PSILs were assessed using transmission electron microscopy, confirming that the PSILs were not aggregated, and have a diameter of less than 120 nm (Fig. 2 b–d). As determined by QELS analyses, the mean diameter of PSLs and PSILs averaged 66 ± 3 nm (Fig. 2e) and 80 ± 4 nm (Fig. 2f), respectively. This suggests that the conjugation of MAbs to PEG strands on the liposomal surface added 14 nm to the liposome diameter. The electron microscopy study (not shown) and QELS size analyses (Fig. 3) demonstrated that the size and morphology of both PSLs and PSILs remained stable. Indeed, no significant change in the mean particle size within the PSL/PSIL suspensions was detectable after up to 21 days when kept in saline buffer at room temperature (Fig. 3a–b). In DMEM, non PEGylated liposomes, PSLs and PSILs remained stable in terms of size for at least 48 h (Fig. 3c), which was longer than the calculated terminal half-life in vivo (see below).
To assess leakage of DNA after the initial synthesis, the amount of [33P]-DNA protected by the liposomes from nuclease digestion was determined at two time points after the first size exclusion chromatography. No significant leakage of [33P]-DNA was measured after 24 and 48 h, indicating that the formulation was stable in HEPES buffer and DMEM (Fig. 4a). Furthermore, the terminal half-life in vivo of [33P]-DNA encapsulated into PSILs was estimated at 23.9 h (Fig. 4b) after a single administration in the tail vein. Although this did not distinguish intact DNA from metabolite, plasmatic [33P]-DNA showed a TCA precipitability of 90% 2 min after the injection.
(a) The % of [33P]-DNA encapsulated into liposomes (L), polyethylene glycol (PEG)-stabilized liposomes (PSLs) and polyethylene glycol (PEG)-stabilized immunoliposomes (PSILs) remained over 75% after storage of the preparation in 0.05 M HEPES buffer or in Dulbecco’s Modified Eagle’s Medium (DMEM), at 37°C for up to 48 h. (b) Determination of the terminal half-life of [33P]-DNA encapsulated into PSILs injected in the tail vein of rats. Values are mean ± SEM of three animals.
Analysis of Encapsulated DNA
To ascertain that the encapsulated DNA was not importantly altered by the encapsulation process, an agarose gel analysis was carried out. Loading of the DNA-PSIL complex caused the majority of encapsulated DNA to remain in the well (lane A4- Fig. 5a), consistent with the incapacity of liposomes to migrate through the agarose matrix. However, a fraction of the encapsulated DNA migrated similarly to control DNA (lane A3- Fig. 5a), probably due to leakage during electrophoresis. Size exclusion chromatography fractions corresponding to digested DNA (see Fig. 1) contained DNA fragments too small to be retained by the agarose gel (lane A5- Fig. 5a). On the other hand, detergent-treated PSILs released all pGL3 plasmid DNA and showed a migration pattern similar to control DNA (lane A6 versus A3—Fig. 5a). Overall, data included in Fig. 5a show that the encapsulation process did not visibly alter the agarose migration pattern of encapsulated DNA.
Agarose gel analysis of DNA plasmids in the encapsulation process. (a) The majority of DNA entrapped into PEG-stabilized immunoliposomes (PSILs) did not migrate freely in the agarose gel (lane A4), whereas detergent-induced lysis of the complex released the DNA (lane A6), which then migrated as control pGL3 plasmid (lane A3). An eluted fraction containing nuclease-digested non-encapsulated DNA plasmid is shown in lane A5. (b) Demonstration that the 40% ethanol (EtOH) concentration used during encapsulation did not alter the subsequent digestion of unencapsulated plasmids. Addition of nucleases (1 h at 37°C) totally digested the pGL3 DNA plasmid (lane B2), with or without 40% EtOH (lanes B3 and B4). To mimic usual experimental conditions, empty liposomes were added to the pGL3 plasmid sample but did not interfere with digestion of unencapsulated DNA (lane B5). (c) Encapsulation efficiencies as measured with liquid scintillation counting (LSC) of eluted fractions following the size exclusion chromatography were comparable to those estimated from UV optical density analysis of agarose gels. Values are mean ± SEM of 58 and three experiments for LSC and gel analyses, respectively.
Exposure of the PSIL preparations to nucleases is an essential step aimed at discarding unencapsulated DNA to obtain an accurate quantification of the DNA complexed within and protected by the liposomes. Thus, it was important to rule out whether the ethanol-based encapsulation procedure affected the efficacy of the nuclease degradation. Agarose gel analysis of DNA digested by nucleases in presence of 40% ethanol clearly shows that ethanol did not affect the catalytic properties of the enzymes in the protocol (lane B4 versus B3 and B2—Fig. 5b). Furthermore, empty liposomes did not interfere with the activity of the nucleases (lane B5—Fig. 5b). In a separate agarose gel analysis of eluted fractions from size exclusion chromatography, it was also estimated by UV optical density analysis that 79% of DNA remained entrapped within the liposomes, similar to percentages determined by liquid scintillation counting analysis of [33P]-DNA (Fig. 5c).
In Vitro Transfection Study
The transferrin receptor-dependent transfection capacity of PSILs was demonstrated in cultured N2A mouse cells (Fig. 6). Significant luciferase expression was observed in wells containing N2A cells transfected with PSILs coupled to RI7 (18-fold increase versus unconjugated PSLs, p < 0.001) or 8D3 (21-fold increase versus unconjugated PSLs, p < 0.001) MAbs raised against the mouse transferrin receptor (Fig. 6). Addition of an excess of free RI7 MAbs competed with available transferrin receptors on N2A cells and blocked the transfection, resulting in luciferase expression levels comparable to unconjugated PSLs (Fig. 6). pGLuc showed higher levels of expressed luciferase than pGL3 or pGL4 in this model (not shown). No signs of toxicity from the PSIL formulation or excess RI7 MAbs were observed in cultured N2A cells (not shown).
Transferrin receptor-specific transfection of neuroblastoma Neuro 2A (N2A) cultured cells with the pGLuc plasmid encapsulated into PEG-stabilized immunoliposomes (PSILs) targeting transferrin receptors. Luciferase expression in N2A cells 48 h after transfection with PSILs conjugated to RI7 or 8D3 monoclonal antibodies (MAbs) and containing 160 ng of pGLuc was significantly higher than unconjugated (NC) PSLs. Addition of free RI7 MAbs (0.5 mg/ml) competed with available transferrin receptors and blocked transfection of N2A cells with PSILs conjugated to either RI7 or 8D3 MAbs. Relative light units (RLU) per g of proteins are expressed as mean ± SD of at least three separate wells. * P < 0.001 versus non-conjugated PSL-pGLuc; † P < 0.001 versus RI7-PSIL-pGLuc; § P < 0.001 versus 8D3-PSIL-pGLuc, using an ANOVA followed by a Tukey’s multiple comparison test.
DISCUSSION
Liposome-based gene delivery combines the advantages of safety and the potential for tissue-selective drug targeting. Furthermore, PSLs can be mass-produced at relatively low cost and have already been approved for clinical use (10–12,25). The present report describes a new methodology to generate small unilamellar neutral PSLs with a high DNA encapsulation efficiency in a reproducible fashion. The PSLs can be routinely conjugated to proteic vectors such as MAbs targeting the mouse transferrin receptor to form PSILs. Encapsulated reporter genes were used to confirm the transferrin receptor-dependent transfection of N2A cells with the present novel PSIL formulation.
The high DNA encapsulation efficiency shown here into a PSIL formulation designed for in vivo applications compares advantageously to previously published methodologies. Indeed, most procedures to generate liposome-DNA complexes of less than 150 nm in diameter using either freeze-thaw cycles, reverse-phase evaporation, or dehydration-rehydration techniques report limited encapsulation efficiencies (17,32,33,38,40,41). For example, a commonly used freeze–thaw technique was associated with encapsulation efficiencies inferior to 20% along with important interassay variations (Table I). More recently, Bailey and Sullivan (2000) were able to significantly improve DNA encapsulation efficiencies by condensing DNA with calcium ions and ethanol during the entrapment process (38). A similar ethanol-based approach suitable for mass production has been recently published (41). However, in its original form, the method of Bailey and Sullivan (2000) produces neutral liposomes between 150 and 200 nm that are not PEG-stabilized nor conjugated to targeting vectors, therefore less suitable for in vivo use.
Conjugation of proteic vectors to liposomes is necessary for target-specific gene delivery. Here, it is shown that thiolation of the MAbs using a 2-iminothiolane : MAb ratio of 40: 1 leads to very high conjugation efficiencies to the PSL-DNA complex along with a 14 nm increase in particle diameter. The number of MAbs attached per liposome is an important factor for cellular uptake. Low MAb : liposome ratios might lead to insufficient target binding, while an excess of MAbs attached to a liposome might interfere with delivery (33,42). The best in vitro transfection efficiency was obtained with a 2-iminothiolane : MAb ratio of 40 : 1, which corresponded to about 92 MAbs per liposome.
An issue often raised to criticize liposome formulations is that their shelf life or their blood circulation time can be limited. QELS analysis and electron microscopy showed that the present liposome preparation is stable at least in terms of size and morphology. Moreover, no DNA leakage or size alteration were detected after 48 h at 37°C in HEPES buffer or DMEM. Thus, the characterization of the size and stability of the PSILs along with the conserved migration pattern of encapsulated DNA are consistent with a drug delivery formulation suitable for in vivo use. Consistently, our pharmacokinetic experiment showed that the terminal half-life of the preparation was over 20 h after a single intravenous injection in rats. Therefore, based on the present data and on many features shared with previously published PSIL formulations successfully used in vivo (17,18,23), it is expected that this novel formulation has most prerequisites for in vivo investigations.
The N2A cell line expresses the transferrin receptor (36,37) and thus represented a very good model to investigate the targeting capabilities of the present liposomal formulation. The transferrin receptor has been shown to transport transferrin-bound iron into the cell through a well described ATP-dependent endocytosis mechanism (43). The data clearly show that the conjugated RI7 or 8D3 MAbs targeting the transferrin receptors on N2A cells induced transfection of these cells. Without the conjugated vector or in presence of an excess of free vectors, no transfection occurred, indicating the selectivity of the vector targeting. Since transferrin receptors are enriched within the brain capillary endothelium, which forms the blood-brain barrier in vivo (43–45), the present formulation will be investigated as a brain-targeting carrier in the mouse.
The need to develop efficient lipid-based transfection vehicles is highlighted by recent clinical trials, which have demonstrated the safety risks associated with viral vectors. Indeed, virtually all humans have a pre-existing immunity to HSV-1, AV and, to a lesser extent, AAV (6,7,10). Hence, intracerebral injections of these vectors can cause rate-limiting inflammation and demyelination. Life-threatening inflammatory responses have been described after injections of HSV and AV vectors in humans and monkeys (6,8). Another major concern is that retrovirus, lentivirus or AAV can randomly and stably integrate into the host genome to produce long-lasting expression of exogenous genes after transfection (6). Random uncontrolled incorporation of an exogenous gene carries a risk of mutagenesis. This aspect has been evidenced by a recent clinical trial with severe combined immunodeficiency (SCID) children in France who received retroviral gene therapy in 2000 and are now being treated for acute T-cell leukemia due to insertion of the retroviral vector near the promoter of the proto-oncogene LMO2 (9,10). In summary, problems with pharmaceutical processing and toxicity associated with viral vectors call for the development of alternative gene therapy vectors.
Although PSILs represent a safe alternative for targeted gene delivery compared to viral vectors, refinement of current methodologies is required. Poor transfection efficiency compared to that obtained with viral vectors is still a major problem, which will likely be solved through the development of powerful expression plasmids with features improving mRNA stability, reducing bacterial DNA content, and inserting endosomal disruption sequences or nuclear-import signals, for example (10,46). The high DNA encapsulation efficiencies reported here offer several advantages such as economy in materials including expression plasmids, proteic vectors, smaller injection volumes and reduced waste. Therefore, the present formulation is one of the first reported to combine many critical characteristics required for efficient targeted drug delivery and high transfection rates in vitro and in vivo.
Abbreviations
- 2I:
-
2-iminothiolane
- AAV:
-
adeno-associated virus
- ATP:
-
adenosine 5′-triphosphate
- AV:
-
adenovirus
- BSA:
-
bovine serum albumine
- CMV:
-
cytomegalovirus
- [33P]-dCTP:
-
deoxycytidine 5′-[α-33P]triphosphate
- DDAB:
-
didodecyldimethylammonium bromide
- Dpm:
-
disintegrations per minute
- DSPE:
-
distearoylphosphatidylethanolamine
- EtOH:
-
ethanol
- HEPES:
-
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- HSV-1:
-
herpes simplex virus-1
- LSC:
-
liquid scintillation counting
- MAbs:
-
monoclonal antibodies
- MWCO:
-
molecular weight cut-off
- N2A:
-
neuroblastoma neuro 2A
- NC:
-
non-conjugated
- 3H-NSP:
-
N-succinimidyl-[2,3-3H] propionate
- PBS:
-
phosphate buffered saline
- PCR:
-
polymerase chain reaction
- pGLuc:
-
pCMV-GLuc
- PEG:
-
polyethylene glycol
- PEG2000 :
-
2,000 da polyethylene glycol
- POPC:
-
1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine
- PSLs:
-
PEG-stabilized liposomes
- PSILs:
-
PEG-stabilized immunoliposomes
- RLU:
-
relative light units
- RT:
-
room temperature
- QELS:
-
quasi-elastic light scattering
- SEM:
-
standard error mean
- SCID:
-
severe combined immunodeficiency
- SD:
-
standard deviation
- SV40:
-
simian virus 40
- VP-SFM:
-
virus production serum-free medium
References
M. G. Kaplitt and D. W. Pfaff. Viral vectors for gene delivery and expression in the CNS. Methods 10:343–350 (1996).
J. H. Kordower, M. E. Emborg, J. Bloch, S. Y. Ma, Y. Chu, L. Leventhal, J. McBride, E. Y. Chen, S. Palfi, B. Z. Roitberg, W. D. Brown, J. E. Holden, R. Pyzalski, M. D. Taylor, P. Carvey, Z. Ling, D. Trono, P. Hantraye, N. Deglon, and P. Aebischer. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson’s disease. Science 290:767–773 (2000).
B. L. Davidson and X. O. Breakefield. Viral vectors for gene delivery to the nervous system. Nat. Rev., Neurosci. 4:353–364 (2003).
P. R. Lowenstein and M. G. Castro. Recent advances in the pharmacology of neurological gene therapy. Curr. Opin. Pharmacol. 4:91–97 (2004).
P. L. Sinn, S. L. Sauter, and P. B. McCray, Jr. Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors—design, biosafety, and production. Gene Ther. 12:1089–1098 (2005).
C. E. Thomas, A. Ehrhardt, and M. A. Kay. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev., Genet. 4:346–358 (2003).
K. Jooss and N. Chirmule. Immunity to adenovirus and adeno-associated viral vectors: implications for gene therapy. Gene Ther. 10:955–963 (2003).
M. A. Schnell, Y. Zhang, J. Tazelaar, G. P. Gao, Q. C. Yu, R. Qian, S. J. Chen, A. N. Varnavski, C. LeClair, S. E. Raper, and J. M. Wilson. Activation of innate immunity in nonhuman primates following intraportal administration of adenoviral vectors. Molec. Ther. 3:708–722 (2001).
S. Hacein-Bey-Abina, C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415–419 (2003).
D. J. Glover, H. J. Lipps, and D. A. Jans. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev., Genet. 6:299–310 (2005).
N. Smyth Templeton. Liposomal delivery of nucleic acids in vivo. DNA Cell Biol. 21:857–867 (2002).
T. M. Allen and P. R. Cullis. Drug delivery systems: entering the mainstream. Science 303:1818–1822 (2004).
D. J. Bharali, I. Klejbor, E. K. Stachowiak, P. Dutta, I. Roy, N. Kaur, E. J. Bergey, P. N. Prasad, and M. K. Stachowiak. Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc. Natl. Acad. Sci. U. S. A. 102:11539–11544 (2005).
P. H. Tan, M. Manunta, N. Ardjomand, S. A. Xue, D. F. Larkin, D. O. Haskard, K. M. Taylor, and A. J. George. Antibody targeted gene transfer to endothelium. J. Gene Med. 5:311–323 (2003).
Y. Zhang, H. Jeong Lee, R. J. Boado, and W. M. Pardridge. Receptor-mediated delivery of an antisense gene to human brain cancer cells. J. Gene Med. 4:183–194 (2002).
P. Machy, F. Lewis, L. McMillan, and Z. L. Jonak. Gene transfer from targeted liposomes to specific lymphoid cells by electroporation. Proc. Natl. Acad. Sci. U. S. A. 85:8027–8031 (1988).
Y. Zhang, F. Calon, C. Zhu, R. J. Boado, and W. M. Pardridge. Intravenous nonviral gene therapy causes normalization of striatal tyrosine hydroxylase and reversal of motor impairment in experimental parkinsonism. Hum. Gene Ther. 14:1–12 (2003).
N. Shi, Y. Zhang, C. Zhu, R. J. Boado, and W. M. Pardridge. Brain-specific expression of an exogenous gene after i.v. administration. Proc. Natl. Acad. Sci. U. S. A. 98:12754–12759 (2001).
C. Y. Wang and L. Huang. pH-sensitive immunoliposomes mediate target-cell-specific delivery and controlled expression of a foreign gene in mouse. Proc. Natl. Acad. Sci. U. S. A. 84:7851–7855 (1987).
N. Zhu, D. Liggitt, Y. Liu, and R. Debs. Systemic gene expression after intravenous DNA delivery into adult mice. Science 261:209–211 (1993).
A. Gabizon, H. Shmeeda, and Y. Barenholz. Pharmacokinetics of pegylated liposomal Doxorubicin: review of animal and human studies. Clin. Pharmacokinet. 42:419–436 (2003).
P. R. Cullis, A. Chonn, and S. C. Semple. Interactions of liposomes and lipid-based carrier systems with blood proteins: relation to clearance behaviour in vivo. Adv. Drug Deliv. Rev. 32:3–17 (1998).
K. Maruyama, O. Ishida, T. Takizawa, and K. Moribe. Possibility of active targeting to tumor tissues with liposomes. Adv. Drug Deliv. Rev. 40:89–102 (1999).
A. L. Klibanov, K. Maruyama, V. P. Torchilin, and L. Huang. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 268:235–237 (1990).
A. Schnyder and J. Huwyler. Drug transport to brain with targeted liposomes. NeuroRx 2:99–107 (2005).
P. Goyal, K. Goyal, S. G. Kumar, A. Singh, O. P. Katare, and D. N. Mishra. Liposomal drug delivery systems—clinical applications. Acta Pharm. 55:1–25 (2005).
N. Shi and W. M. Pardridge. Noninvasive gene targeting to the brain. Proc. Natl. Acad. Sci. U. S. A. 97:7567–7572 (2000).
L. Cattel, M. Ceruti, and F. Dosio. From conventional to stealth liposomes: a new frontier in cancer chemotherapy. Tumori 89:237–249 (2003).
R. I. Mahato, K. Kawabata, Y. Takakura, and M. Hashida. in vivo disposition characteristics of plasmid DNA complexed with cationic liposomes. J. Drug Target. 3:149–157 (1995).
G. Osaka, K. Carey, A. Cuthbertson, P. Godowski, T. Patapoff, A. Ryan, T. Gadek, and J. Mordenti. Pharmacokinetics, tissue distribution, and expression efficiency of plasmid [33P]DNA following intravenous administration of DNA/cationic lipid complexes in mice: use of a novel radionuclide approach. J. Pharm. Sci. 85:612–618 (1996).
L. D. Leserman, P. Machy, and J. Barbet. Cell-specific drug transfer from liposomes bearing monoclonal antibodies. Nature 293:226–228 (1981).
P. A. Monnard, T. Oberholzer, and P. Luisi. Entrapment of nucleic acids in liposomes. Biochim. Biophys. Acta 1329:39–50 (1997).
N. H. Kim, H. M. Park, S. Y. Chung, E. J. Go, and H. J. Lee. Immunoliposomes carrying plasmid DNA: preparation and characterization. Arch. Pharm. Res. 27:1263–1269 (2004).
D. D. Lasic, B. Ceh, M. C. Stuart, L. Guo, P. M. Frederik, and Y. Barenholz. Transmembrane gradient driven phase transitions within vesicles: lessons for drug delivery. Biochim. Biophys. Acta 1239:145–156 (1995).
M. L. Lee, W. Y. Poon, and H. S. Kingdon. A two-phase linear regression model for biologic half-life data. J. Lab. Clin. Med. 115:745–748 (1990).
I. J. Hildebrandt, M. Iyer, E. Wagner, and S. S. Gambhir. Optical imaging of transferrin targeted PEI/DNA complexes in living subjects. Gene Ther. 10:758–764 (2003).
R. R. Nixon. Prion-associated increases in Src-family kinases. J. Biol. Chem. 280:2455–2462 (2005).
A. L. Bailey and S. M. Sullivan. Efficient encapsulation of DNA plasmids in small neutral liposomes induced by ethanol and calcium. Biochim. Biophys. Acta 1468:239–252 (2000).
Y. Fang, T. S. Spisz, and J. H. Hoh. Ethanol-induced structural transitions of DNA on mica. Nucleic Acids Res. 27:1943–1949 (1999).
A. Cudd and C. Nicolau. Intracellular fate of liposome-encapsulated DNA in mouse liver. Analysis using electron microscope autoradiography and subcellular fractionation. Biochim. Biophys. Acta 845:477–491 (1985).
L. B. Jeffs, L. R. Palmer, E. G. Ambegia, C. Giesbrecht, S. Ewanick, and I. MacLachlan. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 22:362–372 (2005).
J. Huwyler, D. Wu, and W. M. Pardridge. Brain drug delivery of small molecules using immunoliposomes. Proc. Natl. Acad. Sci. U. S. A. 93:14164–14169 (1996).
Z. M. Qian, H. Li, H. Sun, and K. Ho. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol Rev. 54:561–587 (2002).
W. A. Jefferies, M. R. Brandon, S. V. Hunt, A. F. Williams, K. C. Gatter, and D. Y. Mason. Transferrin receptor on endothelium of brain capillaries. Nature 312:162–163 (1984).
D. C. Mash, J. Pablo, D. D. Flynn, S. M. Efange, and W. J. Weiner. Characterization and distribution of transferrin receptors in the rat brain. J. Neurochem. 55:1972–1979 (1990).
R. J. Boado and W. M. Pardridge. Ten nucleotide cis element in the 3′-untranslated region of the GLUT1 glucose transporter mRNA increases gene expression via mRNA stabilization. Mol. Brain Res. 59:109–113 (1998).
Acknowledgements
Grants from the Canadian Institutes of Health Research (CIHR) (FC-RMP72549 and M2C-63922), the Alzheimer Society Canada (FC-ASC 0516), and the Parkinson Society Canada (FC 2004) funded this research. VR was supported by FORMSAV-CIHR and Laval University “Fonds d’Enseignement et de Recherche” faculty of pharmacy studentships.
Competing interests statement
The authors declare that they have no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Rivest, Phivilay, contributed equally to this work.
Rights and permissions
About this article
Cite this article
Rivest, V., Phivilay, A., Julien, C. et al. Novel Liposomal Formulation for Targeted Gene Delivery. Pharm Res 24, 981–990 (2007). https://doi.org/10.1007/s11095-006-9224-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-006-9224-x