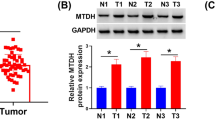
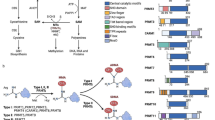
Abstract
Evidence demonstrates that DNA methylation is associated with the occurrence and development of various neurological diseases. However, the potential target genes undergoing DNA methylation, as well as their involvement in the chemotherapy drug oxaliplatin-induced neuropathic pain, are still unclear. Here, Lrfn4, which showed hypermethylation in the promoter regions, was screened from the SRA methylation database (PRJNA587622) following oxaliplatin treatment. MeDIP and qPCR assays identified that oxaliplatin treatment increased the methylation in Lrfn4 promoter region and decreased the expression of LRFN4 in the spinal dorsal horn. The assays with gain and loss of LRFN4 function demonstrated that LRFN4 downregulation in spinal dorsal horn contributed to the oxaliplatin-induced mechanical allodynia and cold hyperalgesia. Moreover, oxaliplatin treatment increased the DNA methyltransferases DNMT3a expression and the interaction between DNMT3a and Lrfn4 promoter, while inhibition of DNMT3a prevented the downregulation of LRFN4a induced by oxaliplatin. We also observed that the transcriptional factor POU2F1 can bind to the predicted sites in DNMT3a promoter region, oxaliplatin treatment upregulated the expression of transcriptional factor POU2F1 in dorsal horn neurons. Intrathecal injection of POU2F1 siRNA prevented the DNMT3a upregulation and the LRFN4 downregulation induced by oxaliplatin. Additionally, intrathecal injection of DNMT3a siRNA or POU2F1 siRNA alleviated the mechanical allodynia induced by oxaliplatin. These findings suggested that transcription factor POU2F1 upregulated the expression of DNMT3a, which subsequently decreased LRFN4 expression through hypermethylation modification in spinal dorsal horn, thereby mediating neuropathic pain following oxaliplatin treatment.
Highlights
Downregulation of LRFN4 mediated by DNA methyltransferases DNMT3a in spinal dorsal horn contributed to the neuropathic pain induced by chemotherapy agent oxaliplatin treatment.
Transcription factor POU2F1 bound to the Dnmt3a promoter and increased the DNMT3a expression in oxliplatin-induced neuropathic pain.





Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Saif MW, Reardon J (2005) Management of oxaliplatin-induced peripheral neuropathy. Ther Clin Risk Manag 1:249–258
Loprinzi CL, Lacchetti C, Bleeker J, Cavaletti G, Chauhan C, Hertz DL, Kelley MR, Lavino A, Lustberg MB, Paice JA, Schneider BP, Lavoie Smith EM, Smith ML, Smith TJ, Wagner-Johnston N, Hershman DL (2020) Prevention and Management of Chemotherapy-Induced Peripheral Neuropathy in Survivors of adult cancers: ASCO Guideline Update. J Clin Oncol 38:3325–3348
Miltenburg NC, Boogerd W (2014) Chemotherapy-induced neuropathy: a comprehensive survey. Cancer Treat Rev 40:872–882
Yuzaki M (2018) Two classes of secreted synaptic organizers in the Central Nervous System. Annu Rev Physiol 80:243–262
Lie E, Li Y, Kim R, Kim E (2018) SALM/Lrfn family synaptic adhesion molecules. Front Mol Neurosci 11:105
Nam J, Mah W, Kim E (2011) The SALM/Lrfn family of leucine-rich repeat-containing cell adhesion molecules. Semin Cell Dev Biol 22:492–498
Morimura N, Inoue T, Katayama K, Aruga J (2006) Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene 380:72–83
Lie E, Yeo Y, Lee EJ, Shin W, Kim K, Han KA, Yang E, Choi TY, Bae M, Lee S, Um SM, Choi SY, Kim H, Ko J, Kim E (2021) SALM4 negatively regulates NMDA receptor function and fear memory consolidation. Commun Biol 4:1138
Li Y, Zhang P, Choi TY, Park SK, Park H, Lee EJ, Lee D, Roh JD, Mah W, Kim R, Kim Y, Kwon H, Bae YC, Choi SY, Craig AM, Kim E (2015) Splicing-dependent trans-synaptic SALM3-LAR-RPTP interactions regulate excitatory Synapse Development and Locomotion. Cell Rep 12:1618–1630
Anis S, Mosek A (2018) [Epigenetic Mechanisms in Models of Chronic Pain - a target for Novel Therapy?]. Harefuah 157:370–373
Chen YY, Jiang KS, Bai XH, Liu M, Lin SY, Xu T, Wei JY, Li D, Xiong YC, Xin WJ, Li ZY (2021) ZEB1 Induces Ddr1 Promoter Hypermethylation and Contributes to the Chronic Pain in Spinal Cord in Rats Following Oxaliplatin Treatment. Neurochem Res
Chen Z, Zhang Y (2020) Role of mammalian DNA methyltransferases in Development. Annu Rev Biochem 89:135–158
Zhao JY, Liang L, Gu X, Li Z, Wu S, Sun L, Atianjoh FE, Feng J, Mo K, Jia S, Lutz BM, Bekker A, Nestler EJ, Tao YX (2017) DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun 8:14712
Zhao FQ (2013) Octamer-binding transcription factors: genomics and functions. Front Biosci (Landmark Ed) 18:1051–1071
Sturm RA, Das G, Herr W (1988) The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev 2:1582–1599
Yuan J, Wen J, Wu S, Mao Y, Mo K, Li Z, Su S, Gu H, Ai Y, Bekker A, Zhang W, Tao YX (2019) Contribution of dorsal root ganglion octamer transcription factor 1 to neuropathic pain after peripheral nerve injury. Pain 160:375–384
Zhang XZ, Luo DX, Bai XH, Ding HH, Liu M, Deng J, Mai JW, Yang YL, Zhang SB, Ruan XC, Zhang XQ, Xin WJ, Xu T (2020) Upregulation of TRPC6 mediated by PAX6 hypomethylation is involved in the mechanical allodyina induced by chemotherapeutics in dorsal root ganglion. Int J Neuropsychopharmacol
Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY (2019) AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 47:D33–D38
Wang J, Zhang XS, Tao R, Zhang J, Liu L, Jiang YH, Ma SH, Song LX, Xia LJ (2017) Upregulation of CX3CL1 mediated by NF-kappaB activation in dorsal root ganglion contributes to peripheral sensitization and chronic pain induced by oxaliplatin administration. Mol Pain 13:1744806917726256
Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38
Turek-Plewa J, Jagodzinski PP (2005) The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 10:631–647
Pance A (2016) Oct-1, to go or not to go? That is the PolII question. Biochim Biophys Acta 1859:820–824
Garriga J, Laumet G, Chen SR, Zhang Y, Madzo J, Issa JJ, Pan HL, Jelinek J (2018) Nerve Injury-Induced Chronic Pain is Associated with persistent DNA methylation reprogramming in dorsal Root Ganglion. J Neurosci 38:6090–6101
Liu H (2019) Synaptic organizers: synaptic adhesion-like molecules (SALMs). Curr Opin Struct Biol 54:59–67
Liang L, Lutz BM, Bekker A, Tao YX (2015) Epigenetic regulation of chronic pain. Epigenomics 7:235–245
Lie E, Ko JS, Choi SY, Roh JD, Cho YS, Noh R, Kim D, Li Y, Kang H, Choi TY, Nam J, Mah W, Lee D, Lee SG, Kim HM, Kim H, Choi SY, Um JW, Kang MG, Bae YC, Ko J, Kim E (2016) SALM4 suppresses excitatory synapse development by cis-inhibiting trans-synaptic SALM3-LAR adhesion. Nat Commun 7:12328
Ouyang J, Chen X, Su S, Li X, Xu X, Yu X, Ke C, Zhu X (2021) Neuroligin1 contributes to Neuropathic Pain by promoting phosphorylation of cofilin in excitatory neurons. Front Mol Neurosci 14:640533
Zhu Y, Yao S, Augustine MM, Xu H, Wang J, Sun J, Broadwater M, Ruff W, Luo L, Zhu G, Tamada K, Chen L (2016) Neuron-specific SALM5 limits inflammation in the CNS via its interaction with HVEM. Sci Adv 2:e1500637
Konakahara S, Saitou M, Hori S, Nakane T, Murai K, Itoh R, Shinsaka A, Kohroki J, Kawakami T, Kajikawa M, Masuho Y (2011) A neuronal transmembrane protein LRFN4 induces monocyte/macrophage migration via actin cytoskeleton reorganization. FEBS Lett 585:2377–2384
Greenberg MVC, Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20:590–607
Gowher H, Jeltsch A (2018) Mammalian DNA methyltransferases: new discoveries and open questions. Biochem Soc Trans 46:1191–1202
Sun L, Gu X, Pan Z, Guo X, Liu J, Atianjoh FE, Wu S, Mo K, Xu B, Liang L, Bekker A, Tao YX (2019) Contribution of DNMT1 to Neuropathic Pain Genesis partially through Epigenetically repressing Kcna2 in primary afferent neurons. J Neurosci 39:6595–6607
Sun L, Zhao JY, Gu X, Liang L, Wu S, Mo K, Feng J, Guo W, Zhang J, Bekker A, Zhao X, Nestler EJ, Tao YX (2017) Nerve injury-induced epigenetic silencing of opioid receptors controlled by DNMT3a in primary afferent neurons. Pain 158:1153–1165
Shao C, Gao Y, Jin D, Xu X, Tan S, Yu H, Zhao Q, Zhao L, Wang W, Wang D (2017) DNMT3a methylation in neuropathic pain. J Pain Res 10:2253–2262
Vazquez-Arreguin K, Tantin D (2016) The Oct1 transcription factor and epithelial malignancies: old protein learns new tricks. Biochim Biophys Acta 1859:792–804
Kim K, Kim N, Lee GR (2016) Transcription factors Oct-1 and GATA-3 cooperatively regulate Th2 cytokine gene expression via the RHS5 within the Th2 Locus Control Region. PLoS ONE 11:e0148576
Micheli L, Parisio C, Lucarini E, Vona A, Toti A, Pacini A, Mello T, Boccella S, Ricciardi F, Maione S, Graziani G, Lacal PM, Failli P, Ghelardini C, Di Cesare Mannelli L (2021) VEGF-A/VEGFR-1 signalling and chemotherapy-induced neuropathic pain: therapeutic potential of a novel anti-VEGFR-1 monoclonal antibody. J Exp Clin Cancer Res 40:320
Acknowledgements
This study was funded by National Natural Science Foundation of China (Grant No. 31970936), Guangdong Basic and Applied Basic Research Foundation (2023A1515030020, 2022A1515012259), Guangzhou Science and Technology Plan Project (202206060004, 202201010988).
Funding
This study was funded by National Natural Science Foundation of China (Grant No. 31970936), Guangdong Basic and Applied Basic Research Foundation (2023A1515030020, 2022A1515012259), Guangzhou Science and Technology Plan Project (202206060004, 202201010988).
Author information
Authors and Affiliations
Contributions
Su- Xia Luo and Wen-Jun Xin contributed to the study conception and design. Data collection and analysis were performed by Yan-Hui Gu and Jing Wang. The first draft of the manuscript was written by Yan-Hui Gu and Wei-Cheng Lu. Material preparation were performed Yong Cheng and Rong Tao. Experiments were performed by Yan-Hui Gu, Jing Wang, Shi-Jia Zhang, Ting Xu and Ke-Wei Zhai. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
These authors have declared that no conflict of interest exists.
Disclosure
This study has never been published elsewhere.
Ethics Approval
The experimental protocols were approved by the Animal Care and Use Committee of Sun Yat-sen University and conducted in strict accordance with the guideline of National Institutes of Health on the animal care and the ethical guideline.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gu, YH., Wang, J., Lu, WC. et al. POU2F1/DNMT3a Pathway Participates in Neuropathic Pain by Hypermethylation-Mediated LRFN4 Downregulation Following Oxaliplatin Treatment. Neurochem Res 48, 3652–3664 (2023). https://doi.org/10.1007/s11064-023-04011-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-023-04011-w