Abstract
Purpose
Medulloblastoma is the most frequent pediatric malignant brain tumor, and is divided into four main subgroups: WNT, SHH, group 3, and group 4. MYCN amplification is an important medulloblastoma prognostic biomarker. We aimed to molecular classify and predict MYCN amplification in a single assay.
Methods
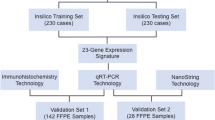
It was included 209 medulloblastomas from 205 patients (Brazil, Argentina, and Portugal), divided into training (n = 50) and validation (n = 159) sets. A nCounter assay was carried out using a custom panel for molecular classification, with additional genes, including MYCN. nSolver 4.0 software and the R environment were used for profiling and MYCN mRNA analysis. MYCN amplification by FISH was performed in 64 cases.
Results
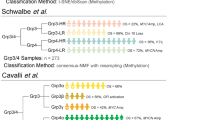
The 205 medulloblastomas were classified in SHH (44.9%), WNT (15.6%), group 3 (18.1%) and group 4 (21.4%). In the training set, MYCN amplification was detected in three SHH medulloblastomas by FISH, which showed significantly higher MYCN mRNA counts than non-FISH amplified cases, and a cutoff for MYCN amplification was established (\(\overline{X }\) + 4σ = 11,124.3). Applying this threshold value in the validation set, we identified MYCN mRNA counts above the cutoff in three cases, which were FISH validated.
Conclusion
We successfully stratified medulloblastoma molecular subgroups and predicted MYCN amplification using a single nCounter assay without the requirement of additional biological tissue, costs, or bench time.





Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACTB:
-
Actin Beta
- CNS:
-
Central nervous system
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- FFPE:
-
Formalin-Fixed Paraffin-Embedded
- FITC:
-
Fluorescein isothiocyanate
- FISH:
-
Fluorescence in situ hybridization
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- HCL:
-
Chloridric acid
- LDH:
-
Lactate Dehydrogenase
- mRNA:
-
Messenger RNA
- MYCN :
-
MYCN Proto-Oncogene
- SD:
-
Standard deviation
- SHH:
-
Sonic hedgehog
- SSC:
-
Saline Sodium Citrate
- WHO:
-
World health organization
- WNT:
-
Wingless
- t-SNE:
-
T-distributed stochastic neighbor embedding
References
Northcott PA, Robinson GW, Kratz CP et al (2019) Medulloblastoma. Nat Rev Dis Primers 5(1):11. https://doi.org/10.1038/s41572-019-0063-6
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Cavalli FMG, Remke M, Rampasek L et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737-754.e6. https://doi.org/10.1016/j.ccell.2017.05.005
Beroukhim R, Mermel CCH, Porter D et al (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463:899–905. https://doi.org/10.1038/NATURE08822
Schaub F, Dhankani V, Berger A et al (2018) Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst 6:282-300.e2. https://doi.org/10.1016/J.CELS.2018.03.003
Matsui A, Ihara T, Suda H et al (2013) Gene amplification: mechanisms and involvement in cancer. Biomol Concepts 4:567–582. https://doi.org/10.1515/BMC-2013-0026
Ruiz-Pérez MV, Henley AB, Arsenian-Henriksson M (2017) The MYCN protein in health and disease. Genes (Basel). https://doi.org/10.3390/GENES8040113
Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD et al (2021) The MYC oncogene—the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol 2021:1–14. https://doi.org/10.1038/s41571-021-00549-2
Hatton BA, Knoepfler PS, Kenney AM et al (2006) N-myc is an essential downstream effector of shh signaling during both normal and neoplastic cerebellar growth. Cancer Res 66:8655–8661. https://doi.org/10.1158/0008-5472.CAN-06-1621
Shrestha S, Morcavallo A, Gorrini C, Chesler L (2021) Biological role of MYCN in medulloblastoma: novel therapeutic opportunities and challenges ahead. Front Oncol. https://doi.org/10.3389/FONC.2021.694320
Pfister S, Remke M, Benner A et al (2009) Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 27:1627–1636. https://doi.org/10.1200/JCO.2008.17.9432
Pfister S, Remke M, Toedt G et al (2007) Supratentorial primitive neuroectodermal tumors of the central nervous system frequently harbor deletions of the CDKN2A locus and other genomic aberrations distinct from medulloblastomas. Genes Chromosom Cancer 46:839–851. https://doi.org/10.1002/gcc.20471
Shakoori AR (2017) Fluorescence in situ hybridization (FISH) and its applications. In: Bhat TA, Wani AA (eds) Chromosom structure and aberrations. Springer, New Delhi, pp 343–367
Pandita A, Bayani J, Paderova J et al (2011) Integrated cytogenetic and high-resolution array CGH analysis of genomic alterations associated with MYCN amplification. Cytogenet Genome Res 134:27–39. https://doi.org/10.1159/000324698
Boensch M, Oberthuer A, Fischer M et al (2005) Quantitative real-time PCR for quick simultaneous determination of therapy-stratifying markers MYCN amplification, deletion 1p and 11q. Diagn Mol Pathol 14:177–182. https://doi.org/10.1097/01.PAS.0000176767.10800.17
Geiss GK, Bumgarner RE, Birditt B et al (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325. https://doi.org/10.1038/NBT1385
Tsang HF, Xue VW, Koh SP et al (2017) NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn 17:95–103. https://doi.org/10.1080/14737159.2017.1268533
Leal LF, Evangelista AF, de Paula FE et al (2018) Reproducibility of the NanoString 22-gene molecular subgroup assay for improved prognostic prediction of medulloblastoma. Neuropathology 38:475–483. https://doi.org/10.1111/neup.12508
da Silva LS, Mançano BM, de Paula FE et al (2020) Expression of GNAS, TP53, and PTEN improves the patient prognostication in sonic hedgehog (SHH) medulloblastoma subgroup. J Mol Diagnostics 22:957–966. https://doi.org/10.1016/j.jmoldx.2020.04.207
Northcott PA, Dubuc AM, Pfister S, Taylor MD (2012) Molecular subgroups of medulloblastoma. Expert Rev Neurother 12:871. https://doi.org/10.1586/ERN.12.66
Novaes L, Sussuchi da Silva L, De Marchi P et al (2021) Simultaneous analysis of Simultaneous analysis of ALK, RET, and ROS1 gene fusions by NanoString in Brazilian lung adenocarcinoma patients. Transl. Lung Cancer Res. 10:292–303. https://doi.org/10.21037/TLCR-20-740
Fuller C, Perry A (2002) Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol 12:67–86. https://doi.org/10.1111/J.1750-3639.2002.TB00424.X
Chrzanowska NM, Kowalewski J, Lewandowska MA (2020) Use of fluorescence in situ hybridization (FISH) in diagnosis and tailored therapies in solid tumors. Molecules. https://doi.org/10.3390/MOLECULES25081864
Aguado C, Teixido C, Román R, Reyes R, Giménez-Capitán A, Marin E, Cabrera C, Viñolas N, Castillo S, Muñoz S, Arcocha A, López-Vilaró L, Sullivan I, Aldeguer E, Rodríguez S, Moya I, Viteri S, Cardona AF, Palmero R, Sainz C, Mesa-Guzmán M, Lozano MD, Aguil RN (2021) Multiplex RNA-based detection of clinically relevant MET alterations in advanced non-small cell lung cancer. Mol Oncol 15:350–363. https://doi.org/10.1002/1878-0261.12861
Korshunov A, Benner A, Remke M et al (2008) Accumulation of genomic aberrations during clinical progression of medulloblastoma. Acta Neuropathol 116:383–390. https://doi.org/10.1007/s00401-008-0422-y
Kumar R, Smith KS, Deng M et al (2021) Clinical outcomes and patient-matched molecular composition of relapsed medulloblastoma. J Clin Oncol 39:807–821. https://doi.org/10.1200/JCO.20.01359
Roussel MF, Robinson GW (2013) Role of MYC in medulloblastoma. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a014308
Borgenvik A, Čančer M, Hutter S et al (2021) Targeting MYCN in molecularly defined malignant brain tumors. Front Oncol. https://doi.org/10.3389/fonc.2020.626751
Goytain A, Ng T (2020) NanoString nCounter technology: high-throughput rna validation. In: Li H, Elfman J (eds) Chimeric RNA: methods and protocols. Springer, New York, NY, pp 125–139
Bracht JWP, Gimenez-Capitan A, Huang CY et al (2021) Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci Rep. https://doi.org/10.1038/S41598-021-83132-0
Souza KCB, Evangelista AF, Leal LF et al (2019) Identification of cell-free circulating microRNAs for the detection of early breast cancer and molecular subtyping. J Oncol. https://doi.org/10.1155/2019/8393769
Causin RL, Da Silva LS, Evangelista AF et al (2021) MicroRNA biomarkers of high-grade cervical intraepithelial neoplasia in liquid biopsy. Biomed Res Int. https://doi.org/10.1155/2021/6650966
Veldman-Jones MH, Brant R, Rooney C et al (2015) Evaluating robustness and sensitivity of the nanostring technologies ncounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res 75:2587–2593. https://doi.org/10.1158/0008-5472.CAN-15-0262
Funding
We want to thank the funding support from Barretos Cancer Hospital and the Public Ministry of Labor (MPT), Campinas (Research, Prevention, and Education of Occupational Cancer, Brazil), and post-doc scholarship for DAM from the National Oncology Care Support Program (PRONON), Brazil. MPT supported LSS.
Author information
Authors and Affiliations
Contributions
DAM (ORCID 0000–0002-0764-447X): Data collection, experimental procedures, data analysis, manuscript writing. LSdS (ORCID 0000–0002-7470–4655): Data analysis, prognostic results discussion, manuscript review. MFZ: FISH experiments, data analysis, and manuscript review. MB: nCounter experiments, manuscript review. FEdP: nCounter experiments, manuscript review. IVVS: Pathological review of tumor samples, results, discussion, manuscript review. GRT: Pathological review of tumor samples, results, discussion, and manuscript review. MdMM: Pathological review of tumor samples, manuscript review. FS: Pathological review of tumor samples, results, discussion, and manuscript review. LNS: Pathological review of tumor samples from Ribeirão Preto Medical School, Ribeirão Preto, São Paulo, Brazil. JNS: Pathological review of tumor samples, results, discussion, and manuscript review. SMFM: Clinical review of cases, results, discussion, and manuscript review. ML: Clinical review of cases, results, discussion, and manuscript review. GNMH: Clinical review of cases, results, discussion, and manuscript review. HG-R (ORCID 0000–0003-2056-464X): Pathological review of tumor samples, results, discussion, and manuscript review. SC: Clinical review of cases, results, discussion, and manuscript review. MJGdC: Clinical review of cases, results, discussion, and manuscript review. SN: Clinical review of cases, results, discussion, and manuscript review. MJS: FISH experiments, data analysis, and manuscript review. JP: Pathological review of tumor samples, results, discussion, and manuscript review. CAJ: Clinical review of cases, results, discussion, and manuscript review. BMM: Clinical review of cases, results, discussion, and manuscript review. RMR (ORCID 0000–0002-9639–7940): Supervisor and project coordinator, results, discussion, manuscript writing, and review.
Corresponding author
Ethics declarations
Conflict of interests
The authors of the present study declare that they have no conflicts of interests.
Ethical approval
The institutional review board approved this retrospective study from Barretos Cancer Hospital (CAAE: 59979816.6.1001.5437 Number: 4.719.466).
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Moreno, D.A., da Silva, L.S., Zanon, M.F. et al. Single nCounter assay for prediction of MYCN amplification and molecular classification of medulloblastomas: a multicentric study. J Neurooncol 157, 27–35 (2022). https://doi.org/10.1007/s11060-022-03965-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-022-03965-1