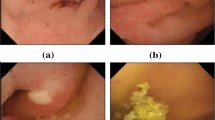
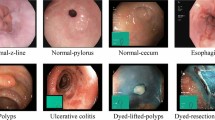
Abstract
Video capsule endoscopy (VCE) is a non-invasive procedure to examine the human bowel. The VCE technology generates thousands of images from different parts of the gastrointestinal tract. Since the examination of these images is a tedious and time-consuming task for doctors, automated diagnosis of digestive diseases from VCE images is highly desired. The majority of the existing studies are based on CNN methods, which are not efficient enough in learning invariant global features in VCE images. Therefore, this paper presents a new framework that combines the learning of global and local features from VCE images. The proposed method utilizes a specific attention mechanism within a convolutional neural network to extract local features, while a vision transformer captures global features. Both local and global features are fused for final classification. Extensive experiments were performed on the public Kvasir Capsule Endoscopy dataset, revealing a promising accuracy of 97%. These results not only highlight the model’s capabilities but also demonstrate its favorable standing when compared to the state-of-the-art methods. Additionally, achieving a recall of 85%, the proposed system demonstrated robust generalization capabilities, performing impressively on an unseen dataset.







Similar content being viewed by others
Data Availibility
The data used in this study are included in the paper and are openly available at https://osf.io/dv2ag/.
References
Organization WH et al (2018) Malnutrition. key facts. World Health Organization, 1–7
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer Journal for Clinicians 68(6):394–424. https://doi.org/10.3322/caac.21492
Iddan G, Meron G, Glukhovsky A, Swain P (2000) Wireless capsule endoscopy. Nature 405(6785):417–417. https://doi.org/10.1038/35013140
Jia X, Xing X, Yuan Y, Xing L, Meng MQ-H (2019) Wireless capsule endoscopy: a new tool for cancer screening in the colon with deep-learning-based polyp recognition. Proceedings of the IEEE 108(1):178–197. https://doi.org/10.1109/JPROC.2019.2950506
Omori T, Hara T, Sakasai S, Kambayashi H, Murasugi S, Ito A, Nakamura S, Tokushige K (2018) Does the pillcam sb3 capsule endoscopy system improve image reading efficiency irrespective of experience? a pilot study. Endoscopy International Open 6(06):669–675. https://doi.org/10.1055/a-0599-5852
Ye Y et al (2013) Bounds on rf cooperative localization for video capsule endoscopy. PhD thesis, Worcester Polytechnic Institute
Lafraxo S, El Ansari M, Koutti L (2023) Computer-aided system for bleeding detection in wce images based on cnn-gru network. Multimedia Tools and Applications 1–26. https://doi.org/10.1007/s11042-023-16305-w
Souaidi M, Lafraxo S, Kerkaou Z, El Ansari M, Koutti L (2023) A multiscale polyp detection approach for gi tract images based on improved densenet and single-shot multibox detector. Diagnostics 13(4):733. https://doi.org/10.3390/diagnostics13040733
Khan MA, Sahar N, Khan WZ, Alhaisoni M, Tariq U, Zayyan MH, Kim YJ, Chang B (2022) Gestronet: a framework of saliency estimation and optimal deep learning features based gastrointestinal diseases detection and classification. Diagnostics 12(11):2718. https://doi.org/10.3390/diagnostics12112718
Dheir IM, Abu-Naser SS (2022) Classification of anomalies in gastrointestinal tract using deep learning
Yuan Y, Li B, Meng MQ-H (2015) Improved bag of feature for automatic polyp detection in wireless capsule endoscopy images. IEEE Trans Auto Sci Eng 13(2):529–535. https://doi.org/10.1109/TASE.2015.2395429
Yu L, Yuen PC, Lai J (2012) Ulcer detection in wireless capsule endoscopy images. In: Proceedings of the 21st international conference on pattern recognition (ICPR2012), pp 45–48. IEEE
Figueiredo IN, Kumar S, Leal C, Figueiredo PN (2013) Computer-assisted bleeding detection in wireless capsule endoscopy images. Comput Methods Biomech Biomed Eng: Imag Visual 1(4):198–210. https://doi.org/10.1080/21681163.2013.796164
Ellahyani A, Jaafari IE, Charfi S, Ansari ME (2021) Detection of abnormalities in wireless capsule endoscopy based on extreme learning machine. Signal Image Video Proc 15(5):877–884. https://doi.org/10.1007/s11760-020-01809-x
Deeba F, Bui FM, Wahid KA (2020) Computer-aided polyp detection based on image enhancement and saliency-based selection. Biomed Signal Proce Control 55:101530. https://doi.org/10.1016/j.bspc.2019.04.007
Souaidi M, Abdelouahed AA, El Ansari M (2019) Multi-scale completed local binary patterns for ulcer detection in wireless capsule endoscopy images. Multimed Tools Appl 78:13091–13108. https://doi.org/10.1007/s11042-018-6086-2
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of the IEEE 86(11):2278–2324. https://doi.org/10.1109/5.726791
Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J (2013) Mitosis detection in breast cancer histology images with deep neural networks. In: International conference on medical image computing and computer-assisted intervention, pp 411–418. https://doi.org/10.1007/978-3-642-40763-5_51 Springer
Garbaz A, Lafraxo S, Charfi S, El Ansari M, Koutti L (2022) Bleeding classification in wireless capsule endoscopy images based on inception-resnet-v2 and cnns. In: 2022 IEEE conference on computational intelligence in bioinformatics and computational biology (CIBCB), pp 1–6. https://doi.org/10.1109/CIBCB55180.2022.9863010 IEEE
Cook D, Feuz KD, Krishnan NC (2013) Transfer learning for activity recognition: a survey. Knowledge and information systems 36(3):537–556. https://doi.org/10.1007/s10115-013-0665-3
Dai Y, Gao Y, Liu F (2021) Transmed: transformers advance multi-modal medical image classification. Diagnostics 11(8):1384. https://doi.org/10.3390/diagnostics11081384
He K, Gan C, Li Z, Rekik I, Yin Z, Ji W, Gao Y, Wang Q, Zhang J, Shen D (2023) Transformers in medical image analysis. Intelligent Medicine 3(1):59–78. https://doi.org/10.1016/j.imed.2022.07.002
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. arXiv:2010.11929
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vision 60:91–110. https://doi.org/10.1023/B:VISI.0000029664.99615.94
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on pattern analysis and machine intelligence 24(7):971–987. https://doi.org/10.1109/TPAMI.2002.1017623
Dalal N, Triggs B (2005) Histograms of oriented gradients for human detection. In: 2005 IEEE computer society conference on computer vision and pattern recognition (CVPR’05), vol 1, pp 886–893. https://doi.org/10.1109/CVPR.2005.177IEEE
Stephane M (1999) A wavelet tour of signal processing. Elsevier. https://doi.org/10.1016/B978-0-12-374370-1.X0001-8
Li B, Meng MQ-H (2012) Automatic polyp detection for wireless capsule endoscopy images. Expert Syst Appl 39(12):10952–10958. https://doi.org/10.1016/j.eswa.2012.03.029
Charfi S, Ansari ME (2018) Computer-aided diagnosis system for colon abnormalities detection in wireless capsule endoscopy images. Multimed Tools Appl 77(3):4047–4064. https://doi.org/10.1007/s11042-017-4555-7
Sainju S, Bui FM, Wahid K (2013) Bleeding detection in wireless capsule endoscopy based on color features from histogram probability. In: 2013 26th IEEE Canadian conference on electrical and computer engineering (CCECE), pp 1–4. https://doi.org/10.1109/CCECE.2013.6567779 . IEEE
Xing X, Jia X, Meng MQ-H (2018) Bleeding detection in wireless capsule endoscopy image video using superpixel-color histogram and a subspace knn classifier. In: 2018 40th Annual international conference of the ieee engineering in medicine and biology society (EMBC), pp 1–4. https://doi.org/10.1109/EMBC.2018.8513012IEEE
Boser BE, Guyon IM, Vapnik VN (1992) A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on computational learning theory, pp 144–152 . https://doi.org/10.1145/130385.130401
Zhu R, Zhang R, Xue D (2015) Lesion detection of endoscopy images based on convolutional neural network features. In: 2015 8th International congress on image and signal processing (CISP), pp 372–376. https://doi.org/10.1109/CISP.2015.7407907 IEEE
Sekuboyina AK, Devarakonda ST, Seelamantula CS (2017) A convolutional neural network approach for abnormality detection in wireless capsule endoscopy. In: 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017), pp 1057–1060. https://doi.org/10.1109/ISBI.2017.7950698 IEEE
Yu J-s, Chen J, Xiang Z, Zou Y-X (2015) A hybrid convolutional neural networks with extreme learning machine for wce image classification. In: 2015 IEEE international conference on robotics and biomimetics (ROBIO), pp 1822–1827. https://doi.org/10.1109/ROBIO.2015.7419037 IEEE
Seguí S, Drozdzal M, Pascual G, Radeva P, Malagelada C, Azpiroz F, Vitrià J (2016) Generic feature learning for wireless capsule endoscopy analysis. Comput Biol Med 79:163–172. https://doi.org/10.1016/j.compbiomed.2016.10.011
Iakovidis DK, Georgakopoulos SV, Vasilakakis M, Koulaouzidis A, Plagianakos VP (2018) Detecting and locating gastrointestinal anomalies using deep learning and iterative cluster unification. IEEE Transactions on Medical Imaging 37(10):2196–2210. https://doi.org/10.1109/TMI.2018.2837002
Goel N, Kaur S, Gunjan D, Mahapatra S (2022) Dilated cnn for abnormality detection in wireless capsule endoscopy images. Soft Comput 26(3):1231–1247. https://doi.org/10.1007/s00500-021-06546-y
Yuan Y, Meng MQ-H (2017) Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 44(4):1379–1389. https://doi.org/10.1002/mp.12147
Khan MA, Khan MA, Ahmed F, Mittal M, Goyal LM, Hemanth DJ, Satapathy SC (2020) Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recogn Lett 131:193–204. https://doi.org/10.1016/j.patrec.2019.12.024
Sharif M, Attique Khan M, Rashid M, Yasmin M, Afza F, Tanik UJ (2021) Deep cnn and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images. J Experim Theor Artif Intell 33(4):577–599. https://doi.org/10.1080/0952813X.2019.1572657
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Caroppo A, Leone A, Siciliano P (2021) Deep transfer learning approaches for bleeding detection in endoscopy images. Comput Med Imag Graphics 88:101852. https://doi.org/10.1016/j.compmedimag.2020.101852
Oukdach Y, Kerkaou Z, El Ansari M, Koutti L, El Ouafdi AF (2022) Gastrointestinal diseases classification based on deep learning and transfer learning mechanism. In: 2022 9th international conference on wireless networks and mobile communications (WINCOM), pp 1–6. https://doi.org/10.1109/WINCOM55661.2022.9966474 IEEE
Souaidi M, El Ansari M (2022) A new automated polyp detection network mp-fssd in wce and colonoscopy images based fusion single shot multibox detector and transfer learning. IEEE Access 10:47124–47140. https://doi.org/10.1109/ACCESS.2022.3171238
Zheng H, Chen H, Huang J, Li X, Han X, Yao J (2019) Polyp tracking in video colonoscopy using optical flow with an on-the-fly trained cnn. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 79–82. https://doi.org/10.1109/ISBI.2019.8759180 IEEE
Jain S, Seal A, Ojha A, Yazidi A, Bures J, Tacheci I, Krejcar O (2021) A deep cnn model for anomaly detection and localization in wireless capsule endoscopy images. Comput Biol Med 137:104789. https://doi.org/10.1016/j.compbiomed.2021.104789
Lafraxo S, Souaidi M, El Ansari M, Koutti L (2023) Semantic segmentation of digestive abnormalities from wce images by using attresu-net architecture. Life 13(3):719. https://doi.org/10.3390/life13030719
Iqbal I, Walayat K, Kakar MU, Ma J (2022) Automated identification of human gastrointestinal tract abnormalities based on deep convolutional neural network with endoscopic images. Intell Syst Appl 16:200149. https://doi.org/10.1016/j.iswa.2022.200149
Lima DLS, Pessoa ACP, De Paiva AC, Silva Cunha AMT, Júnior GB, De Almeida JDS (2022) Classification of video capsule endoscopy images using visual transformers. In: 2022 IEEE-EMBS international conference on biomedical and health informatics (BHI), pp 1–4. https://doi.org/10.1109/BHI56158.2022.9926791 IEEE
Zhang Y, Liu H, Hu Q (2021) Transfuse: fusing transformers and cnns for medical image segmentation. In: Medical image computing and computer assisted intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24, pp 14–24. https://doi.org/10.1007/978-3-030-87193-2_2 Springer
Lin A, Chen B, Xu J, Zhang Z, Lu G, Zhang D (2022) Ds-transunet: dual swin transformer u-net for medical image segmentation. IEEE Trans Inst Measure 71:1–15. https://doi.org/10.1109/TIM.2022.3178991
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Proc Syst, 30. arXiv:1706.03762
Smedsrud PH, Thambawita V, Hicks SA, Gjestang H, Nedrejord OO, Næss E, Borgli H, Jha D, Berstad TJD, Eskeland SL et al (2021) Kvasir-capsule, a video capsule endoscopy dataset. Sci Data 8(1):142. https://doi.org/10.1038/s41597-021-00920-z
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778. arXiv:1512.03385
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258. arXiv:1610.02357
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708. arXiv:1608.06993
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 8697–8710. arXiv:1707.07012
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence, vol 31. https://doi.org/10.1609/aaai.v31i1.11231
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning, pp 6105–6114. arXiv:1905.11946 PMLR
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626. arXiv:1610.02391
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826. arXiv:1512.00567
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C (2018) Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520. arXiv:1801.04381
Jain S, Seal A, Ojha A (2022) A hybrid convolutional neural network with meta feature learning for abnormality detection in wireless capsule endoscopy images. arXiv:2207.09769
Jain S, Seal A, Ojha A, Krejcar O, Bureš J, Tachecí I, Yazidi A (2020) Detection of abnormality in wireless capsule endoscopy images using fractal features. Computers in biology and medicine 127:104094. https://doi.org/10.1016/j.compbiomed.2020.104094
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F (2015) Wm-dova maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graphics 43:99–111. https://doi.org/10.1016/j.compmedimag.2015.02.007
Funding
This work was supported by the Ministry of National Education by Vocational Training; in part by the Higher Education and Scientific Research through the Ministry of Industry, Trade, and Green and Digital Economy; in part by the Digital Development Agency (ADD); and in part by the National Center for Scientific and Technical Research (CNRST) under Project ALKHAWARIZMI/2020/20.
Author information
Authors and Affiliations
Contributions
Y.O., Z.K., M.E., L.K., and A.F.E. wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oukdach, Y., Kerkaou, Z., El Ansari, M. et al. ViTCA-Net: a framework for disease detection in video capsule endoscopy images using a vision transformer and convolutional neural network with a specific attention mechanism. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-023-18039-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-023-18039-1