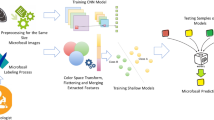
Abstract
Paleontologists generally use a low-cost electro-optical system to classify microfossils. This manual identification is a time-consuming process and it may take about a long time, especially if there are thousands of microfossil samples. In order to solve this problem, we propose a hybrid method based on Convolutional Neural Networks (CNN) and Bidirectional/Long Short-Time Memory (LSTM/BiLSTM) networks for the automatic classification of Globotruncana microfossil species. First, the images of microfossil samples were collected with a low-cost system and labeled by a paleontologist. After preprocessing, the classification is carried out with different combinations of CNN, LSTM, and Bidirectional LSTM (BiLSTM) models from the scratch developed in this paper. Finally, detailed experimental analyses have been made using accuracy, sensitivity, specificity, precision, F-score, and area under curve metrics. In the existing literature, as far as we know, this study is the first investigation work of prediction Globotruncana microfossil species using hybrid deep learning algorithms. Experiments demonstrate that the proposed models have reached the best accuracy with 97.35% and the best AUC score of 0.968 for automatic identification of Globotruncana microfossil species.





















Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ali A, Zhu Y, Chen Q, et al (2019) Leveraging Spatio-temporal patterns for predicting citywide traffic crowd flows using deep hybrid neural networks. In: 2019 IEEE 25th international conference on parallel and distributed systems (ICPADS). IEEE, pp 125–132
Ali A, Zhu Y, Zakarya M (2021) A data aggregation based approach to exploit dynamic spatio-temporal correlations for citywide crowd flows prediction in fog computing. Multimed Tools Appl 80:31401–31433. https://doi.org/10.1007/s11042-020-10486-4
Anderson TI, Vega B, Kovscek AR (2020) Multimodal imaging and machine learning to enhance microscope images of shale. Comput Geosci 145:104593. https://doi.org/10.1016/j.cageo.2020.104593
Bhadoria R (2019) The biometric computing: recognition and registration. CRC Press
Bolli HM (1957) The genera Praeglobotruncana, Rotalipora, Globotruncana and Abathomphalus in the upper Cretaceous of Trinidad, BWI. US Natl Museum Bull 215:51–60
Bollmann J, Quinn PS, Vela M, Brabec B, Brechner S, Cortés MY, Hilbrecht H, Schmidt DN, Schiebel R, Thierstein HR (2005) Automated particle analysis: calcareous microfossils. In: Image Analysis, Sediments and Paleoenvironments. Kluwer Academic Publishers, Dordrecht, pp. 229–252
Brönnimann P (1952) Globigerinidae from the upper cretaceous Cenomanian-Maestrichtian of Trinidad, BWI. Paleontological Research Institution, Ithaca, N.Y
Bronnimann P, Brown NK (1956) Taxonomy of the Globotruncanidae. Eclogae Geol Helv 48:503–562
Brotzen F (1942) Die Foraminiferengattung Gavelinella nov. gen. und die Systematik der Rotaliiformes. Kungl. boktryckeriet P.A. Norstedt
Cai L, Gao J, Zhao D (2020) A review of the application of deep learning in medical image classification and segmentation. Ann Transl Med 8:713–713. https://doi.org/10.21037/atm.2020.02.44
Caron M (1985) Cretaceous Planktic foraminifera. In: Bolli HM, Saunders JB, Perch Nielsen K (eds) Plankton stratigraphy. Cambridge University Press, Cambridge, pp 17–86
Carvalho LE, Fauth G, Baecker Fauth S, Krahl G, Moreira AC, Fernandes CP, von Wangenheim A (2020) Automated microfossil identification and segmentation using a deep learning approach. Mar Micropaleontol 158:101890. https://doi.org/10.1016/j.marmicro.2020.101890
Cetin O (2022) Accent recognition using a spectrogram image feature-based convolutional neural network. Arab J Sci Eng. https://doi.org/10.1007/s13369-022-07086-9
Charles JJ (2011) Automatic recognition of complete palynomorphs in digital images. Mach Vis Appl 22:53–60. https://doi.org/10.1007/s00138-009-0200-4
Chen C, Hua Z, Zhang R, Liu G, Wen W (2020) Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed Signal Process Control 57:101819. https://doi.org/10.1016/j.bspc.2019.101819
Cushman JA (1927) An outline of a reclassification of the foraminifera. Contrib from Cushman Lab Foraminifer Res 3:1–105
Díez-Pastor JF, Latorre-Carmona P, Arnaiz-González Á, Ruiz-Pérez J, Zurro D (2020) You are not my type: an evaluation of classification methods for automatic Phytolith identification. Microsc Microanal 26:1158–1167. https://doi.org/10.1017/S1431927620024629
Elder LE, Hsiang AY, Nelson K, Strotz LC, Kahanamoku SS, Hull PM (2018) Data descriptor: sixty-one thousand recent planktonic foraminifera from the Atlantic Ocean. Sci Data 5:1–12. https://doi.org/10.1038/sdata.2018.109
Encyclopædia Britannica (2017) Paleontology science
Gorur K, Bozkurt M, Bascil M, Temurtas F (2019) GKP signal processing using deep CNN and SVM for tongue-machine Interface. Trait du Signal 36:319–329. https://doi.org/10.18280/ts.360404
Gorur K, Kaya Ozer C, Ozer I, Can Karaca A, Cetin O, Kocak I (2022) Species-level microfossil prediction for Globotruncana genus using machine learning models. Arab J Sci Eng. https://doi.org/10.1007/s13369-022-06822-5
Harlan Johnson J (1944) Paleontology, petroleum, and the search for oil. Am Assoc Pet Geol Bull 28:902–908. https://doi.org/10.1306/3d93368e-16b1-11d7-8645000102c1865d
Hossin M, Sulaiman M (2015) A review on evaluation metrics for data classification evaluations. Int J Data Min Knowl Manag Process 5:01–11. https://doi.org/10.5121/ijdkp.2015.5201
Hou Y, Cui X, Canul-Ku M, Jin S, Hasimoto-Beltran R, Guo Q, Zhu M (2020) ADMorph: a 3D digital microfossil morphology dataset for deep learning. IEEE Access 8:148744–148756. https://doi.org/10.1109/ACCESS.2020.3016267
Hou Y, Canul-Ku M, Cui X, Hasimoto-Beltran R, Zhu M (2021) Semantic segmentation of vertebrate microfossils from computed tomography data using a deep learning approach. J Micropalaeontol 40:163–173. https://doi.org/10.5194/jm-40-163-2021
Hsiang AY, Brombacher A, Rillo MC, Mleneck-Vautravers MJ, Conn S, Lordsmith S, Jentzen A, Henehan MJ, Metcalfe B, Fenton IS, Wade BS, Fox L, Meilland J, Davis CV, Baranowski U, Groeneveld J, Edgar KM, Movellan A, Aze T, … Hull PM (2019) Endless Forams: >34,000 modern planktonic foraminiferal images for taxonomic training and automated species recognition using convolutional neural networks. Paleoceanogr Paleoclimatology 34:1157–1177. https://doi.org/10.1029/2019PA003612
Huber BT, MacLeod KG, Tur NA (2008) Chronostratigraphic framework for upper Campanian-Maastrichtian sediments on the Blake nose (subtropical North Atlantic). J Foraminifer Res 38:162–182. https://doi.org/10.2113/gsjfr.38.2.162
Islam MZ, Islam MM, Asraf A (2020) A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inf Med Unlocked 20:100412. https://doi.org/10.1016/j.imu.2020.100412
Itaki T, Taira Y, Kuwamori N, Maebayashi T, Takeshima S, Toya K (2020) Automated collection of single species of microfossils using a deep learning–micromanipulator system. Prog Earth Planet Sci 7:19. https://doi.org/10.1186/s40645-020-00332-4
Itaki T, Taira Y, Kuwamori N, Saito H, Ikehara M, Hoshino T (2020) Innovative microfossil (radiolarian) analysis using a system for automated image collection and AI-based classification of species. Sci Rep 10:21136. https://doi.org/10.1038/s41598-020-77812-6
Johansen TH, Sørensen SA (2020) Towards detection and classification of microscopic foraminifera using transfer learning. Proc North Light Deep Learn Work 1:6. https://doi.org/10.7557/18.5144
Karaderi T, Burghardt T, Hsiang AY, et al (2022) Visual microfossil identification via deep metric learning. In: Lecture Notes in Computer Science. pp. 34–46
Keçeli AS, Kaya A, Keçeli SU (2017) Classification of radiolarian images with hand-crafted and deep features. Comput Geosci 109:67–74. https://doi.org/10.1016/j.cageo.2017.08.011
Ketin İ, Gümüş A (1963) Sinop – Ayancık güneyinde üçüncü bölgeye dahil sahaların jeolojisi hakkında rapor (2. kısım : Jura ve Kretase formasyonlarının etüdü).Report No. 288. Ankara
Laporte LF (1988) What, after all, is paleontology? Palaios 3:453
Li J, Li X, He D (2019) A directed acyclic graph network combined with CNN and LSTM for remaining useful life prediction. IEEE Access 7:75464–75475. https://doi.org/10.1109/ACCESS.2019.2919566
Liu X, Jiang S, Wu R, et al (2022) Automatic taxonomic identification based on the fossil image dataset (>415,000 images) and deep convolutional neural networks. Paleobiology 1–22. https://doi.org/10.1017/pab.2022.14
Marchant R, Tetard M, Pratiwi A, Adebayo M, de Garidel-Thoron T (2020) Automated analysis of foraminifera fossil records by image classification using a convolutional neural network. J Micropalaeontol 39:183–202. https://doi.org/10.5194/jm-39-183-2020
Marmo R, Amodio S, Cantoni V (2006) Microfossils shape classification using a set of width values. Proc - Int Conf Pattern Recognit 1:691–694. https://doi.org/10.1109/ICPR.2006.797
Metz CE (1978) Basic principles of ROC analysis. Semin Nucl Med 8:283–298. https://doi.org/10.1016/S0001-2998(78)80014-2
Mitra R, Marchitto TM, Ge Q, Zhong B, Kanakiya B, Cook MS, Fehrenbacher JS, Ortiz JD, Tripati A, Lobaton E (2019) Automated species-level identification of planktic foraminifera using convolutional neural networks, with comparison to human performance. Mar Micropaleontol 147:16–24. https://doi.org/10.1016/j.marmicro.2019.01.005
Oh SL, Ng EYK, Tan RS, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med 102:278–287. https://doi.org/10.1016/j.compbiomed.2018.06.002
Ostrom JH (2020) Stratigraphy and paleontology of the Cloverly formation (lower cretaceous) of the Bighorn basin area, Wyoming and Montana. Yale University Press
Ozer I (2020) Uzun Kısa Dönem Bellek Ağlarını Kullanarak Erken Aşama Diyabet Tahmini. Mühendislik Bilim ve Araştırmaları Derg 2:50–57. https://doi.org/10.46387/bjesr.790225
Ozer I, Cetin O, Gorur K, Temurtas F (2021) Improved machine learning performances with transfer learning to predicting need for hospitalization in arboviral infections against the small dataset. Neural Comput & Applic 33:14975–14989. https://doi.org/10.1007/S00521-021-06133-0/TABLES/7
Ozer I, Efe SB, Ozbay H (2021) A combined deep learning application for short term load forecasting. Alex Eng J 60:3807–3818. https://doi.org/10.1016/j.aej.2021.02.050
Pedraza A, Bueno G, Deniz O, Cristóbal G, Blanco S, Borrego-Ramos M (2017) Automated diatom classification (part B): a deep learning approach. Appl Sci 7:460. https://doi.org/10.3390/app7050460
Pessagno EA (1967) Upper cretaceous planktonic foraminifera from the Western gulf coastal plain. Paleontological Research Institution
Petrizzo MR (2002) Palaeoceanographic and palaeoclimatic inferences from late cretaceous planktonic foraminiferal assemblages from the Exmouth plateau (ODP sites 762 and 763, eastern Indian Ocean). Mar Micropaleontol 45:117–150. https://doi.org/10.1016/S0377-8398(02)00020-8
Petrizzo MR, Falzoni F, Silva IP (2011) Identification of the base of the lower-to-middle Campanian Globotruncana ventricosa zone: comments on reliability and global correlations. Cretac Res 32:387–405. https://doi.org/10.1016/j.cretres.2011.01.010
Pires de Lima R, Bonar A, Coronado DD et al (2019) Deep convolutional neural networks as a geological image classification tool. Sediment Rec 17:4–9. https://doi.org/10.2110/sedred.2019.2.4
Pires De Lima R, Welch KF, Barrick JE et al (2020) Convolutional neural networks as an aid to biostratigraphy and micropaleontology: a test on late Paleozoic microfossils. Palaios 35:391–402. https://doi.org/10.2110/palo.2019.102
Postuma JA (1971) Manual of planktonic foraminifera. 1–406
Prothero DR (2007) Evolution: what the fossils say and why it matters. Colombia University Press
Rafi SH, Nahid-Al-Masood DSR, Hossain E (2021) A short-term load forecasting method using integrated CNN and LSTM network. IEEE Access 9:32436–32448. https://doi.org/10.1109/ACCESS.2021.3060654
Rawat W, Zenghu W (2018) Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput 29:2352–2449. https://doi.org/10.1162/NECO
Rehn E, Rehn A, Possemiers A (2019) Fossil charcoal particle identification and classification by two convolutional neural networks. Quat Sci Rev 226:106038. https://doi.org/10.1016/j.quascirev.2019.106038
Reichel M (1950) Observations sur les Globotruncana du gisement de la Breggia (Tessin). Eclogae Geol Helv 42:596–617
Renaudie J, Gray R, Lazarus D (2018) Accuracy of a neural net classification of closely-related species of microfossils from a sparse dataset of unedited images. 0–15. https://doi.org/10.7287/peerj.preprints.27328v1
Rhanoui M, Mikram M, Yousfi S, Barzali S (2019) A CNN-BiLSTM model for document-level sentiment analysis. Mach Learn Knowl Extr 1:832–847. https://doi.org/10.3390/make1030048
Robaszynski F, Foraminifera. EWG on P, (France) M de la géologie (1984) Atlas of late cretaceous Globotruncanids. The Group, Paris
Singh LK, Pooja GH et al (2021) An enhanced deep image model for glaucoma diagnosis using feature-based detection in retinal fundus. Med Biol Eng Comput 59:333–353. https://doi.org/10.1007/s11517-020-02307-5
Solano GA, Gasmen P, Marquez EJ (2019) Radiolarian classification decision support using supervised and unsupervised learning approaches. 2018 9th Int Conf information, Intell Syst Appl IISA 2018. https://doi.org/10.1109/IISA.2018.8633617
Waikato TU of (2021) https://sci.waikato.ac.nz/evolution/fossils.shtml. Accessed 07. 08. 2022
Xiang L, Wang P, Yang X, Hu A, Su H (2021) Fault detection of wind turbine based on SCADA data analysis using CNN and LSTM with attention mechanism. Measurement 175:109094. https://doi.org/10.1016/j.measurement.2021.109094
Xiao S, Laflamme M (2009) On the eve of animal radiation: phylogeny, ecology and evolution of the Ediacara biota. Trends Ecol Evol 24:31–40. https://doi.org/10.1016/j.tree.2008.07.015
Xu Y, Dai Z, Wang J, Li Y, Wang H (2020) Automatic recognition of palaeobios images under microscope based on machine learning. IEEE Access 8:172972–172981. https://doi.org/10.1109/ACCESS.2020.3024819
Yang S, Berdine G (2017) The receiver operating characteristic (ROC) curve. Southwest Respir Crit Care Chronicles 5:34. https://doi.org/10.12746/swrccc.v5i19.391
Yasuhara M, Huang H-H, Hull P, et al (2020) Time machine biology: cross-timescale integration of ecology, evolution, and oceanography. Oceanography 33:. https://doi.org/10.5670/oceanog.2020.225
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ozer, I., Ozer, C.K., Karaca, A.C. et al. Species-level microfossil identification for globotruncana genus using hybrid deep learning algorithms from the scratch via a low-cost light microscope imaging. Multimed Tools Appl 82, 13689–13718 (2023). https://doi.org/10.1007/s11042-022-13810-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13810-2