Abstract
Autoinduction systems in Escherichia coli can control the production of proteins without the addition of a particular inducer. In the present study, we optimized the heterologous expression of Moloney Murine Leukemia Virus derived Reverse Transcriptase (MMLV-RT) in E. coli. Among 4 autoinduction media, media Imperial College resulted the highest MMLV-RT overexpression in E. coli BL21 Star (DE3) with incubation time 96 h. The enzyme was produced most optimum in soluble fraction of lysate cells. The MMLV-RT was then purified using the Immobilized Metal Affinity Chromatography method and had specific activity of 629.4 U/mg. The system resulted lower specific activity and longer incubation of the enzyme than a classical Isopropyl ß-D-1-thiogalactopyranoside (IPTG)-induction system. However, the autoinduction resulted higher yield of the enzyme than the conventional induction (27.8%). Techno Economic Analysis revealed that this method could produce MMLV-RT using autoinduction at half the cost of MMLV-RT production by IPTG-induction. Bioprocessing techniques are necessary to conduct to obtain higher quality of MMLV-RT under autoinduction system.






Similar content being viewed by others
Data availability
Data will be made available on request.
References
Haddad F, Baldwin KM (2010) Reverse transcription of the ribonucleic acid: the first step in RT-PCR assay. Methods Mol Biol. https://doi.org/10.1007/978-1-60761-629-0_17
Rifkind D, Freeman GL (2005) Reverse transcriptase. The Nobel Prize winning discoveries in infectious diseases. Elsevier, pp 115–117
Kostoulas P, Eusebi P, Hartnack S (2021) diagnostic accuracy estimates for COVID-19 real-time polymerase chain reaction and lateral flow immunoassay tests with bayesian latent-class models. Am J Epidemiol 190:1689–1695. https://doi.org/10.1093/aje/kwab093
Saeed H, Osama H, Madney YM, Harb HS, Abdelrahman MA, Ehrhardt C, Abdelrahim MEA (2021) COVID-19; current situation and recommended interventions. Int J Clin Pract. https://doi.org/10.1111/ijcp.13886
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172
Fakruddin Md, Bin MKS, Andrews S (2013) Viable but nonculturable bacteria: food safety and public health perspective. ISRN Microbiol 2013:1–6. https://doi.org/10.1155/2013/703813
Kotewicz ML, D’Alessio JM, Driftmier KM, Blodgett KP, Gerard GF (1985) Cloning and overexpression of moloney murine leukemia virus reverse transcriptase in Escherichia coli. Gene 35:249–258. https://doi.org/10.1016/0378-1119(85)90003-4
Tanese N, Roth M, Goff SP (1985) Expression of enzymatically active reverse transcriptase in Escherichia coli. Proc Natl Acad Sci 82:4944–4948. https://doi.org/10.1073/pnas.82.15.4944
Zhang Z, Kuipers G, Niemiec Ł, Baumgarten T, Slotboom DJ, de Gier J-W, Hjelm A (2015) High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG. Microb Cell Fact 14:142
Khani M-H, Bagheri M (2020) Skimmed milk as an alternative for IPTG in induction of recombinant protein expression. Protein Expr Purif 170:105593. https://doi.org/10.1016/j.pep.2020.105593
Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. https://doi.org/10.1016/j.pep.2005.01.016
Ukkonen K, Mayer S, Vasala A, Neubauer P (2013) Use of slow glucose feeding as supporting carbon source in lactose autoinduction medium improves the robustness of protein expression at different aeration conditions. Protein Expr Purif 91:147–154. https://doi.org/10.1016/j.pep.2013.07.016
Li Z, Kessler W, Van Den Heuvel J, Rinas U (2011) Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl Microbiol Biotechnol 91:1203–1213
Gordon E, Horsefield R, Swarts HGP, de Pont JJHHM, Neutze R, Snijder A (2008) Effective high-throughput overproduction of membrane proteins in Escherichia coli. Protein Expr Purif 62:1–8. https://doi.org/10.1016/j.pep.2008.07.005
Mayer S, Junne S, Ukkonen K, Glazyrina J, Glauche F, Neubauer P, Vasala A (2014) Lactose autoinduction with enzymatic glucose release: Characterization of the cultivation system in bioreactor. Protein Expr Purif 94:67–72. https://doi.org/10.1016/j.pep.2013.10.024
Potter R, Rosenthal K (2013) High fidelity reverse transcriptase and the uses thereof. US Patent 8,541,219 B2 24 Sept 2013.
Champoux JJ, Schultz SJ (2009) Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. https://doi.org/10.1111/j.1742-4658.2009.06909
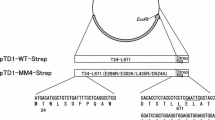
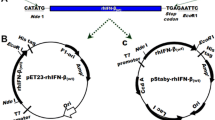
Nuryana I, Laksmi FA, Agustriana E, Dewi KS, Andriani A, Thontowi A, Kusharyoto W, Lisdiyanti P (2022) Expression of codon-optimized gene encoding murine moloney leukemia virus reverse transcriptase in Escherichia coli. Protein J. https://doi.org/10.1007/s10930-022-10066-5
Dewi KS, Permadi DG, Aminah FAM (2020) Expression of recombinant human granulocyte-colony stimulating factor in Escherichia coli using various induction methods. IOP Conf Ser Earth Environ Sci. https://doi.org/10.1088/1755-1315/439/1/012042
Jeong H, Kim HJ, Lee SJ (2015) Complete genome sequence of Escherichia coli strain BL21. Genome Announc. https://doi.org/10.1128/genomeA.00134-15
Du F, Liu Y-Q, Xu Y-S, Li Z-J, Wang Y-Z, Zhang Z-X, Sun X-M (2021) Regulating the T7 RNA polymerase expression in E. coli BL21 (DE3) to provide more host options for recombinant protein production. Microb Cell Fact 20:189
Galluccio M, Console L, Pochini L, Scalise M, Giangregorio N, Indiveri C (2022) Strategies for successful over-expression of human membrane transport systems using bacterial hosts: future perspectives. Int J Mol Sci 23:3823. https://doi.org/10.3390/ijms23073823
Kang T-G, Hong S-H, Jeon G-B, Yang Y-H, Kim S-K (2021) Perturbation of the peptidoglycan network and utilization of the signal recognition particle-dependent pathway enhances the extracellular production of a truncational mutant of CelA in Escherichia coli. J Ind Microbiol Biotechnol. https://doi.org/10.1093/jimb/kuab032
Briand L, Marcion G, Kriznik A, Heydel JM, Artur Y, Garrido C, Seigneuric R, Neiers F (2016) A self-inducible heterologous protein expression system in Escherichia coli. Sci Rep 6:33037. https://doi.org/10.1038/srep33037
Mostovenko E, Deelder AM, Palmblad M (2011) Protein expression dynamics during Escherichia coli glucose-lactose diauxie. BMC Microbiol 11:126. https://doi.org/10.1186/1471-2180-11-126
Traxler MF, Chang D-E, Conway T (2006) Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci 103:2374–2379. https://doi.org/10.1073/pnas.0510995103
Hoopes JT, Elberson MA, Preston RJ, Reddy PT, Kelman Z (2015) Protein labeling in Escherichia coli with 2H, 13C, and 15N. Methods Enzymol. https://doi.org/10.1016/bs.mie.2015.08.023
Yan J (2004) Effects of lactose as an inducer on expression of Helicobacter pylori rUreB and rHpaA, and Escherichia coli rLTKA63 and rLTB. World J Gastroenterol 10:1755. https://doi.org/10.3748/wjg.v10.i12.1755
Fox BG, Blommel PG (2009) Autoinduction of protein expression. Curr Protoc Protein Sci. https://doi.org/10.1002/0471140864.ps0523s56
Rajan R, Ahmed S, Sharma N, Kumar N, Debas A, Matsumura K (2021) Review of the current state of protein aggregation inhibition from a materials chemistry perspective: special focus on polymeric materials. Mater Adv 2:1139–1176. https://doi.org/10.1039/D0MA00760A
Tsumoto K, Ejima D, Kumagai I, Arakawa T (2003) Practical considerations in refolding proteins from inclusion bodies. Protein Expr Purif 28:1–8. https://doi.org/10.1016/S1046-5928(02)00641-1
Mitraki A, King J (1989) Protein folding intermediates and inclusion body formation. Nat Biotechnol 7:690–697. https://doi.org/10.1038/nbt0789-690
Huang C-J, Peng H-L, Patel AK, Singhania RR, Dong C-D, Cheng C-Y (2021) Effects of lower temperature on expression and biochemical characteristics of hcv ns3 antigen recombinant protein. Catalysts 11:1297. https://doi.org/10.3390/catal11111297
Wingfield PT (2014) Preparation of soluble proteins from Escherichia coli. Curr Protoc Protein Sci. https://doi.org/10.1002/0471140864.ps0602s78
Wei D, Wang M, Wang H, Liu G, Fang J, Jiang Y (2022) Development of a method for fast assessment of protein solubility based on ultrasonic dispersion and differential centrifugation technology. ACS Omega 7:31338–31347. https://doi.org/10.1021/acsomega.2c03666
Silprasit K, Thammaporn R, Hannongbua S, Choowongkomon K (2008) Cloning expression purification determining activity of recombinant HIV-1 reverse transcriptase. Agri Nat Resour 42(5):231–239
Lu M, Ngo W, Mei Y, Munshi V, Burlein C, Loughran MH, Williams PD, Hazuda DJ, Miller MD, Grobler JA, Diamond TL, Lai MT (2010) Purification of untagged HIV-1 reverse transcriptase by affinity chromatography. Protein Expr Purif 71(2):231–239. https://doi.org/10.1016/j.pep.2010.01.001
Roshanak S, Yarabbi H, Shahidi F et al (2023) Effects of adding poly-histidine tag on stability, antimicrobial activity and safety of recombinant buforin I expressed in periplasmic space of Escherichia coli. Sci Rep 13:5508–5511. https://doi.org/10.1038/s41598-023-32782-3
Hamdane D, Skouloubris S, Myllykallio H, Golinelli-Pimpaneau B (2010) Expression and purification of untagged and histidine-tagged folate-dependent tRNA:m5U54 methyltransferase from Bacillus subtilis. Protein Expr Purif 73(1):83–89. https://doi.org/10.1016/j.pep.2010.04.013
Fan Q, Neubauer P, Gimpel M (2021) Production of soluble regulatory hydrogenase from Ralstonia eutropha in Escherichia coli using a fed-batch-based autoinduction system. Microb Cell Fact 20:201. https://doi.org/10.1186/s12934-021-01690-4
Tahara N, Tachibana I, Takeo K, Yamashita S, Shimada A, Hashimoto M, Ohno S, Yokogawa T, Nakagawa T, Suzuki F, Ebihara A (2021) Boosting auto-induction of recombinant proteins in Escherichia coli with glucose and lactose additives. Protein Pept Lett 28(10):1180–1190. https://doi.org/10.2174/0929866528666210805120715
Li Z, Kessler W, van den Heuvel J, Rinas U (2011) Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl Microbiol Biotechnol 91(4):1203–1213. https://doi.org/10.1007/s00253-011-3407-z
Chen WB, Nie Y, Mu XQ et al (2014) Auto-induction-based rapid evaluation of extracellular enzyme expression from lac operator-involved recombinant Escherichia coli. Appl Biochem Biotechnol 174:2516–2526. https://doi.org/10.1007/s12010-014-1201-y
Isakova A, Artykov A, Vorontsova Y et al (2023) Application of an autoinduction strategy to optimize the heterologous production of an antitumor bispecific fusion protein based on the TRAIL receptor-selective mutant variant in Escherichia coli. Mol Biotechnol 65:581–589. https://doi.org/10.1007/s12033-022-00561-6
Menacho-Melgar R, Ye Z, Moreb EA, Yang T, Efromson JP, Decker JS, Wang R, Lynch MD (2020) Scalable, two-stage, autoinduction of recombinant protein expression in E. coli utilizing phosphate depletion. Biotechnol Bioeng 117(9):2715–2727
Acknowledgements
The authors would like to thank the Research Team of Extremophile Enzyme, Research Center of Applied Microbiology, National Research and Innovation Agency for permission, guidance, and facilities.
Funding
This research was supported by Indonesia Endowment Fund for Education (LPDP) Program in fiscal year 2021 for National Priority Program for COVID-19.
Author information
Authors and Affiliations
Contributions
CV carried out the research, analyzed and interpreted the data, and wrote the manuscript, FAL designed the research and reviewed the manuscript, IN analyzed and interpreted the data and wrote the manuscript, AA provided supervision, wrote and reviewed the manuscript, NR provided supervision and reviewed the manuscript, EA wrote the manuscript, AP reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors report no conflict of interest.
Ethical approval
It is not applicable for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Handayani, C.V., Laksmi, F.A., Andriani, A. et al. Expression of soluble moloney murine leukemia virus-reverse transcriptase in Escherichia coli BL21 star (DE3) using autoinduction system. Mol Biol Rep 51, 628 (2024). https://doi.org/10.1007/s11033-024-09583-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09583-6