Abstract
Background
Ocimum tenuiflorum L. is a highly traded medicinal with several therapeutic values. Green Tulsi and purple Tulsi are two subtypes in O. tenuiflorum and both have the same medicinal properties. Recent reports have revealed that purple Tulsi contains higher quantities of methyl eugenol (ME), which is moderately toxic and potentially carcinogenic. Therefore, we developed an allele-specific PCR (AS-PCR) method to distinguish the green and purple Tulsi.
Methods and result
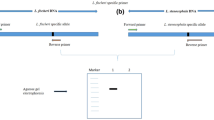
Using the green Tulsi as a reference, 12 single nucleotide polymorphisms (SNPs) and 10 insertions/deletions (InDels) were identified in the chloroplast genome of the purple Tulsi. The C > T SNP at the 1,26,029 position in the ycf1 gene was selected for the development of the AS-PCR method. The primers were designed to amplify 521 bp and 291 bp fragments specific to green and purple Tulsi, respectively. This AS-PCR method was validated in 10 accessions from each subtype and subsequently verified using Sanger sequencing. Subsequently, 30 Tulsi powder samples collected from the market were subjected to molecular identification by AS-PCR. The results showed that 80% of the samples were purple Tulsi, and only 3.5% were green Tulsi. About 10% of the samples were a mixture of both green and purple Tulsi. Two samples (6.5%) did not contain O. tenuiflorum and were identified as O. gratissimum.
Conclusion
The market samples of Tulsi were predominantly derived from purple Tulsi. The AS-PCR method will be helpful for quality control and market surveillance of Tulsi herbal powders.






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References
Ravikumar K, Nooruunisa SB, Ved DK, Bhatt JR, Goraya GS (2018) Compendium of traded Indian Medicinal plants. Foundation for Revitalization of Local Health Traditions (FRLHT), Bengaluru, Karnataka, India
Rastogi S, Kalra A, Gupta V, Khan F, Lal RK, Tripathi AK, Parameswaran S, Gopalakrishnan C, Ramaswamy G, Shasany AK (2015) Unravelling the genome of Holy Basil: an incomparable elixir of life of traditional Indian Medicine. BMC Genomics 16(1):413. https://doi.org/10.1186/s12864-015-1640-z
Kothari SK, Bhattacharya AK, Ramesh S, Garg SN, Khanuja SPS (2005) Volatile constituents in oil from different plant parts of methyl eugenol-rich Ocimum tenuiflorum L.f. (syn. O. Sanctum L.) grown in South India. J Essent Oil Res 17(6):656–658. https://doi.org/10.1080/10412905.2005.9699025
Cohen M (2014) Tulsi - Ocimum sanctum: a herb for all reasons. J Ayurveda Integr Med 5(4):251–259. https://doi.org/10.4103/0975-9476.146554
Jamshidi N, Cohen MM (2017) The clinical efficacy and safety of tulsi in humans: a systematic review of the literature. Evidence-Based Complement Altern Med eCAM 9217567. https://doi.org/10.1155/2017/9217567
Yang L, Zhao F, Zeng B (2016) Electrochemical determination of eugenol using a three-dimensional molecularly imprinted poly (P-aminothiophenol-co-p-aminobenzoic acids) film modified electrode. Electrochim Acta 210:293–300. https://doi.org/10.1016/j.electacta.2016.05.167
Bhuvaneshwari K, Gokulanathan A, Jayanthi M, Govindasamy V, Milella L, Lee S, Yang DC, Girija S (2016) Can Ocimum basilicum L. and Ocimum tenuiflorum L. in vitro culture be a potential source of secondary metabolites? Food Chem 194:55–60. https://doi.org/10.1016/j.foodchem.2015.07.136
Shukla RP, Prasad VG (1985) Population fluctuations of the oriental fruit fly, Dacus dorsalis Hendel in relation to hosts and abiotic factors. Trop Pest Manage 31:273–275. https://doi.org/10.1080/09670878509370999
Ghosh PK, Bhattacharjee P (2016) Mathematical modeling of supercritical carbon dioxide extraction of methyl eugenol from tuberose flowers. Korean J Chem Eng 33:1681–1691. https://doi.org/10.1007/s11814-015-0247-z
European Commission (2001) Opinion of the scientific committee on food on Methyleugenol (4-Allyl-1,2-dimethoxybenzene). URL https://food.ec.europa.eu/system/files/2016-10/fs_food-improvement-agents_flavourings-out102.pdf (Accessed 30.08.2023)
Waddell WJ (2005) Comparisons of thresholds for carcinogenicity on linear and logarithmic dosage scales. Hum Exp Toxicol 24:325–332. https://doi.org/10.1191/0960327105ht525oa
Telci I, Bayram E, Yılmaz G, Avcı B (2006) Variability in essential oil composition of Turkish basils (Ocimum basilicum L). Biochem Syst Ecol 34:489–497. https://doi.org/10.1016/j.bse.2006.01.009
Al-Subeihi AA, Alhusainy W, Kiwamoto R, Spenkelink B, van Bladeren PJ, Rietjens IM, Punt A (2015) Evaluation of the inter individual human variation in bio activation of methyl eugenol using physiologically based kinetic modelling and Monte Carlo simulations. Toxicol Appl Pharmcol 283(2):117–126. https://doi.org/10.1016/j.taap.2014.12.009
WHO (2008) Evaluation of certain food additives and contaminants: sixty-ninth report of the joint FAO/WHO expert committee on food additives. World Health Organ URL https://apps.who.int/iris/handle/10665/44062 (Accessed 5.3.23)
SCCNFP (2000) European Commission Scientific Committees: Expert panel on effective ways of investing in health. Opinion concerning Methyleugenol adopted by the SCCNFP during the 14th plenary meeting of 24 October 2000 | Scientific Committees. URL https://ec.europa.eu/health/scientific_committees/consumer_safety/opinions/sccnfp_opinions_97_04/sccp_out126_en.htm (Accessed 5.4.23)
Health Canada (2010) List of Prohibited and Restricted Cosmetic Ingredients (The Cosmetic Ingredient Hotlist). Canada.ca. URL https://www.canada.ca/en/health-canada/services/consumer-product-safety/cosmetics/cosmetic-ingredient-hotlist-prohibited-restricted-ingredients/changes.html#june2010 (Accessed 5.4.23)
Kitchlu S, Bhadauria R, Ram G, Bindu K, Khajuria RK, Ahuja A (2013) Chemo-divergence in essential oil composition among thirty one core collections of Ocimum sanctum L. grown under Sub-tropical Region of Jammu, India. Am J Plant Sci 04:302–308. https://doi.org/10.4236/ajps.2013.42040
Mondello L, Zappia G, Cotroneo A, Bonaccorsi I, Chowdhury JU, Yusuf M, Dugo G (2002) Studies on the essential oil-bearing plants of Bangladesh. Part VIII. Composition of some Ocimum oils O. basilicum L. var. purpurascens; O. sanctum L. green; O. sanctum L. purple; O. americanum L., citral type; O. americanum L., camphor type. Flavour and Fragrance Journal 17:335–340. https://doi.org/10.1002/ffj.1108
Rana VS, Blazquez MA (2015) Essential oil composition of the aerial parts of five Ocimum species from Western India. J Essent Oil Bearing Plants 18:1234–1241. https://doi.org/10.1080/0972060X.2014.931255
Christina VLP, Annamalai A (2014) Nucleotide based validation of Ocimum species by evaluating three candidate barcodes of the chloroplast region. Mol Ecol Resour 14:60–68. https://doi.org/10.1111/1755-0998.12167
Balaji R, Parani M (2022) DNA barcoding of the market samples of single-drug Herbal Powders reveals adulteration with Taxonomically unrelated plant species. Diversity 14(6):495. https://doi.org/10.3390/d14060495
Ríos-Rodríguez D, Sahi VP, Nick P (2021) Authentication of holy basil using markers relating to a toxicology-relevant compound. Eur Food Res Technol 247:2485–2497. https://doi.org/10.1007/s00217-021-03812-z
Park MJ, Kim MK, In JG, Yang DC (2006) Molecular identification of Korean ginseng by amplification refractory mutation system-PCR. Food Res Int 39:568–574. https://doi.org/10.1016/j.foodres.2005.11.004
In JG, Kim MK, Lee OR, Kim YJ, Lee BS, Kim SY, Kwon WS, Yang DC (2010) Molecular Identification of Korean Mountain Ginseng using an amplification refractory mutation system (ARMS). J Ginseng Res 34(1):41–46. https://doi.org/10.5142/JGR.2010.34.1.041
Li G, Chen Y, Wang R, Wang H, Wang Y (2020) Variety origin authentication of Panax ginseng C.A. Mey. And industrial ginseng products using SNP-based allele-specific PCR method. J Appl Res Med Aromatic Plants 18:100258. https://doi.org/10.1016/j.jarmap.2020.100258
Choi SJ, Ramekar RV, Kim YB, Kim SW, Noh HS, Lee JK, Park NI, Choi IY, Choi SK, Park KC (2017) Molecular authentication of two medicinal plants Ligularia Fischeri and Ligularia Stenocephala using allele-specific PCR (AS-PCR) strategy. Genes Genomics 39:913–920. https://doi.org/10.1007/s13258-017-0554-3
Harini P, Balaji R, Parani M (2021) The complete chloroplast genome of Ocimum tenuiflorum L. subtype Rama Tulsi and its phylogenetic analysis. Mitochondrial DNA Part B 6(8):2224–2226. https://doi.org/10.1080/23802359.2021.1944381
Kavya NM, Balaji R, Tanuja, Parani M, Senthilkumar P (2021) The complete chloroplast genome of Ocimum tenuiflorum L. subtype Krishna Tulsi and its phylogenetic analysis. Mitochondrial DNA Part B 6(8):2358–2360. https://doi.org/10.1080/23802359.2021.1951133
Tamura K, Stecher G, Kumar S (2021) Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120. MEGA11: Molecular Evolutionary Genetics Analysis Version 11
Wangkumhang P, Chaichoompu K, Ngamphiw C, Ruangrit U, Chanprasert J, Assawamakin A, Tongsima S (2007) WASP: a web-based allele-specific PCR assay designing tool for detecting SNPs and mutations. BMC Genomics 8:275. https://doi.org/10.1186/1471-2164-8-275
Awasthi PK, Dixit SC (2007) Chemical compositions of Ocimum sanctum Shyama and Ocimum sanctum Rama Oils from the plains of Northern India. J Essent Oil Bearing Plants 10(4):292–296. https://doi.org/10.1080/0972060X.2007.10643557
Raina AP, Misra RC (2018) Chemo-divergence in essential oil composition among germplasm collection of five Ocimum species from eastern coastal plains of India. J Essent Oil Res 30(1):47–55. https://doi.org/10.1080/10412905.2017.1371087
Johnson JD, Ryan MJ, Toft JD II, Graves SW, Hejtmancik MR, Cunningham ML, Herbert R, Abdo KM (2000) Two-year toxicity and carcinogenicity study of methyleugenol in F344/N rats and B6C3F1 mice. J Agric Food Chem 48(8):3620–3632. https://doi.org/10.1021/jf000364a
Williams GM, Iatropoulos MJ, Jeffrey AM, Duan JD (2013) Methyleugenol hepatocellular cancer initiating effects in rat liver. Food Chem Toxicol 53. https://doi.org/10.1016/j.fct.2012.11.050. :187 – 96
Kool A, de Boer HJ, Kruger A, Rydberg A, Abbad A, Bjork L, Martin G (2012) Molecular identification of commercialized medicinal plants in southern Morocco. PLoS ONE 7:e39459. https://doi.org/10.1371/journal.pone.0039459
Vassou SL, Kusuma G, Parani M (2015) DNA barcoding for species identification from dried and powdered plant parts: a case study with authentication of the raw drug market samples of Sida cordifolia. Gene 559(1):86–93. https://doi.org/10.1016/j.gene.2015.01.025
Shanmughanandhan D, Ragupathy S, Newmaster SG, Mohanasundaram S, Sathishkumar R (2016) Estimating Herbal Product Authentication and Adulteration in India using a Vouchered, DNA-Based Biological Reference Material Library. Drug Saf 39(12):1211–1227. https://doi.org/10.1007/s40264-016-0459-0
Nithaniyal S, Vassou SL, Poovitha S, Balaji R, Parani M (2017) Identification of species adulteration in traded medicinal plant raw drugs using DNA barcoding. Genome 60(2):139–146. https://doi.org/10.1139/gen-2015-0225
Urumarudappa SKJ, Tungphatthong C, Sukrong S (2019) Mitigating the impact of admixtures in Thai Herbal products. Front Pharmacol 10:01205. https://doi.org/10.3389/fphar.2019.01205
Amritha N, Bhooma V, Parani M (2020) Authentication of the market samples of Ashwagandha by DNA barcoding reveals that powders are significantly more adulterated than roots. J Ethnopharmacol 256:112725. https://doi.org/10.1016/j.jep.2020.112725
Theodoridis S, Stefanaki A, Tezcan M, Aki C, Kokkini S, Vlachonasios KE (2012) DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çeşme-Karaburun Peninsula (Turkey). Mol Ecol Resour 12(4):620–633. https://doi.org/10.1111/j.1755-0998.2012.03129.x
Jurges G, Sahi V, Rodriguez DR, Reich E, Bhamra S, Howard C, Slater A, Nick P (2018) Product authenticity versus globalization-the Tulsi case. PLoS ONE 13:e0207763. https://doi.org/10.1371/journal.pone.0207763
Drescher A, Ruf S, Calsa JC, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22(2):97–104. https://doi.org/10.1046/j.1365-313x.2000.00722.x
Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7(4):e35071. https://doi.org/10.1371/journal.pone.0035071
Schinle T, Merfort I, Frommenwiler D, Reich E (2014) Identification of Chemotypesin Holy Basil (Ocimum Sanctum) by High Performance Thin Layer Chromatography (HPTLC). Planta Med 80:P2B18. https://doi.org/10.1055/s-0034-1394895.
Acknowledgements
The authors acknowledge the SRM Institute of Science and Technology.
Funding
This work was supported by the SRM-DBT Partnership Platform for Contemporary Research Services and Skill Development in Advanced Life Sciences Technologies [Grant Number BT/PR12987/INF/22/205/2015].
Author information
Authors and Affiliations
Contributions
R.B: Data curation, Formal analysis, Methodology, Visualization, Writing - original draft. M.P: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing - review & editing. All authors have reviewed the final version of the manuscript and approved it for publication.
Corresponding author
Ethics declarations
Ethical approval
No animal testing was performed during this study. Ocimum tenuiflorum L. is not a protected or endangered species. Sampling activities were not performed at locations where specific permission is required.
Consent to participate
The study does not involve any animal participants and human subjects; this is not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Balaji, R., Parani, M. Development of an allele-specific PCR (AS-PCR) method for identifying high-methyl eugenol-containing purple Tulsi (Ocimum tenuiflorum L.) in market samples. Mol Biol Rep 51, 439 (2024). https://doi.org/10.1007/s11033-024-09365-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09365-0