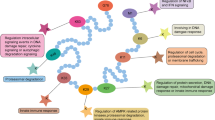
Abstract
Autophagy is a process that is characterized by the destruction of redundant components and the removal of dysfunctional ones to maintain cellular homeostasis. Autophagy dysregulation has been linked to various illnesses, such as neurodegenerative disorders and cancer. The precise transcription of the genes involved in autophagy is regulated by a network of epigenetic factors. This includes histone modifications and histone-modifying enzymes. Epigenetics is a broad category of heritable, reversible changes in gene expression that do not include changes to DNA sequences, such as chromatin remodeling, histone modifications, and DNA methylation. In addition to affecting the genes that are involved in autophagy, the epigenetic machinery can also alter the signals that control this process. In cancer, autophagy plays a dual role by preventing the development of tumors on one hand and this process may suppress tumor progression. This may be the control of an oncogene that prevents autophagy while, conversely, tumor suppression may promote it. The development of new therapeutic strategies for autophagy-related disorders could be initiated by gaining a deeper understanding of its intricate regulatory framework. There is evidence showing that certain machineries and regulators of autophagy are affected by post-translational and epigenetic modifications, which can lead to alterations in the levels of autophagy and these changes can then trigger disease or affect the therapeutic efficacy of drugs. The goal of this review is to identify the regulatory pathways associated with post-translational and epigenetic modifications of different proteins in autophagy which may be the therapeutic targets shortly.
Key Points
1. This review aims to determine the association between the post-translational modulation of autophagy proteins and the regulatory pathways.
2. This review identifies the impact of a variety of autophagic inhibitors and epigenetic drugs on the maintenance and development of the process of autophagy thus revealing the role of these autophagic regulators in cancer therapy.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- CMA:
-
Chaperone mediated autophagy
- ELP3:
-
Elongator complex protein 3
- GBM:
-
Glioblastoma multiforme
- HAT:
-
Histone acetyltransferase
- HDAC:
-
Histone deacetylase
- LAMP:
-
Lysosome-associated membrane protein
- MPP:
-
Mitochondrial Processing Peptidase
- mTORC1:
-
Mammalian target of rapamycin complex 1
- NBR1:
-
Neighbor of BRCA1 gene 1
- PAK1:
-
p21-activated kinase 1
- PAS:
-
Phagophore assembly site
- PE:
-
Phosphatidyl ethanolamine
- PTM:
-
Post-translational modification
- SNAP29:
-
Synaptosome associated protein 29
- TFEB:
-
Transcription factor EB
- TOPK:
-
T-LAK cell‐originated protein kinase
- TRAF6:
-
Tumor necrosis factor receptor-associated factor 6
- UVRAG:
-
UV radiation resistance associated
- VAMP8:
-
Vesicle associated membrane protein 8
- ZBTB16:
-
Zinc finger and BTB domain-containing protein 16
References
Lim SM, Mohamad Hanif EA, Chin SF (2021) Is targeting autophagy mechanism in cancer a good approach? The possible double-edge sword effect. Cell Biosci 11(1):56. https://doi.org/10.1186/s13578-021-00570-z
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20(3):460–473. https://doi.org/10.1089/ars.2013.5371
Chowdhury SG, Bhattacharya D, Karmakar P (2022) Exosomal long noncoding RNAs - the lead thespian behind the regulation, cause and cure of autophagy-related Diseases. Mol Biol Rep 49(7):7013–7024. https://doi.org/10.1007/s11033-022-07514-x
Feng Y, He D, Yao Z, Klionsky DJ (2014) The machinery of macroautophagy. Cell Res 24(1):24–41. https://doi.org/10.1038/cr.2013.168
Oku M, Sakai Y (2018) Three distinct types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. BioEssays 40(6):e1800008. https://doi.org/10.1002/bies.201800008
Bejarano E, Cuervo AM (2010) Chaperone-mediated autophagy. Proc Am Thorac Soc 7(1):29–39. https://doi.org/10.1513/pats.200909-102JS
Guo F, Liu X, Cai H, Le W (2018) Autophagy in neurodegenerative Diseases: pathogenesis and therapy. Brain Pathol 28(1):3–13. https://doi.org/10.1111/bpa.12545
Shu F, Xiao H, Li QN et al (2023) Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct Target Ther 8(1):32. https://doi.org/10.1038/s41392-022-01300-8
Yun CW, Lee SH (2018) The roles of Autophagy in Cancer. Int J Mol Sci 19(11):3466. https://doi.org/10.3390/ijms19113466
George MD, Baba M, Scott SV, Mizushima N et al (2000) Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell 11(3):969–982. https://doi.org/10.1091/mbc.11.3.969
Orsi A, Razi M, Dooley HC, Robinson D et al (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23(10):1860–1873. https://doi.org/10.1091/mbc.E11-09-0746
Takahara T, Amemiya Y, Sugiyama R, Maki M, Shibata H (2020) Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes. J Biomed Sci 27(1):87. https://doi.org/10.1186/s12929-020-00679-2
Noda NN, Fujioka Y (2015) Atg1 family kinases in autophagy initiation. Cell Mol Life Sci 72(16):3083–3096. https://doi.org/10.1007/s00018-015-1917-z
Park JM, Jung CH, Seo M, Otto NM et al (2016) The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 12(3):547–564. https://doi.org/10.1080/15548627.2016.1140293
Hu Y, Reggiori F (2022) Molecular regulation of autophagosome formation. Biochem Soc Trans 50(1):55–69. https://doi.org/10.1042/BST20210819
Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H (2018) The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A 115(41):10363–10368. https://doi.org/10.1073/pnas.1806727115
Hollenstein DM, Kraft C (2020) Autophagosomes are formed at a distinct cellular structure. Curr Opin Cell Biol 65:50–57. https://doi.org/10.1016/j.ceb.2020.02.012
Geng J, Klionsky DJ (2008) The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9(9):859–864. https://doi.org/10.1038/embor.2008.163
Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F (2013) Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep 14(2):206–211. https://doi.org/10.1038/embor.2012.208
Martens S, Fracchiolla D (2020) Activation and targeting of ATG8 protein lipidation. Cell Discov 6:23. https://doi.org/10.1038/s41421-020-0155-1
Tanida I, Ueno T, Kominami E (2008) LC3 and Autophagy. Methods Mol Biol 445:77–88. https://doi.org/10.1007/978-1-59745-157-4_4
Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14(2):207–215. https://doi.org/10.1080/15548627.2017.1378838
Runwal G, Stamatakou E, Siddiqi FH, Puri C et al (2019) LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep 9(1):10147. https://doi.org/10.1038/s41598-019-46657-z
Yang Z, Huang J, Geng J, Nair U, Klionsky DJ (2006) Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 17(12):5094–5104. https://doi.org/10.1091/mbc.e06-06-0479
Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (review). Int J Mol Med 40(2):271–280. https://doi.org/10.3892/ijmm.2017.3036
Ghosh Chowdhury S, Ray R, Karmakar P (2023) Relating aging and autophagy: a new perspective towards the welfare of human health. EXCLI J 22:732–748. https://doi.org/10.17179/excli2023-6300
Schreiber A, Collins BC, Davis C et al (2021) Multilayered regulation of autophagy by the Atg1 kinase orchestrates spatial and temporal control of autophagosome formation. Mol Cell 81(24):5066–5081e10. https://doi.org/10.1016/j.molcel.2021.10.024
Kraft C, Kijanska M, Kalie E et al (2012) Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J 31(18):3691–3703. https://doi.org/10.1038/emboj.2012.225
Keil E, Höcker R, Schuster M et al (2013) Phosphorylation of Atg5 by the Gadd45β-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ 20(2):321–332. https://doi.org/10.1038/cdd.2012.129
Feng X, Zhang H, Meng L et al (2021) Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy 17(3):723–742. https://doi.org/10.1080/15548627.2020.1731266
Hosokawa N, Hara T, Kaizuka T et al (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20(7):1981–1991. https://doi.org/10.1091/mbc.e08-12-1248
Zachari M, Ganley IG (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61(6):585–596. https://doi.org/10.1042/EBC20170021
Kim J, Kundu M, Viollet B, Guan KL (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13(2):132–141. https://doi.org/10.1038/ncb2152
Lu H, Xiao J, Ke C et al (2019) TOPK inhibits autophagy by phosphorylating ULK1 and promotes glioma resistance to TMZ. Cell Death Dis 10(8):583. https://doi.org/10.1038/s41419-019-1805-9
Fujiwara N, Usui T, Ohama T, Sato K (2016) Regulation of Beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3). J Biol Chem 291(20):10858–10866. https://doi.org/10.1074/jbc.M115.704908
McKnight NC, Zhenyu Y (2013) Beclin 1, an essential component and Master Regulator of PI3K-III in Health and Disease. Curr Pathobiol Rep 1(4):231–238. https://doi.org/10.1007/s40139-013-0028-5
Lee B, Park SJ, Lee S et al (2022) Lomitapide, a cholesterol-lowering drug, is an anticancer agent that induces autophagic cell death via inhibiting mTOR. Cell Death Dis 13(7):603. https://doi.org/10.1038/s41419-022-05039-6
Dibble CC, Cantley LC (2015) Regulation of mTORC1 by PI3K signaling. Trends Cell Biol 25(9):545–555. https://doi.org/10.1016/j.tcb.2015.06.002
Di Malta C, Cinque L, Settembre C (2019) Transcriptional regulation of Autophagy: mechanisms and Diseases. Front Cell Dev Biol 7:114. https://doi.org/10.3389/fcell.2019.00114
Cui Z, Napolitano G, de Araujo MEG et al (2023) Structure of the lysosomal mTORC1-TFEB-Rag-ragulator megacomplex. Nature 614(7948):572–579. https://doi.org/10.1038/s41586-022-05652-7
Dossou AS, Basu A (2019) The emerging roles of mTORC1 in Macromanaging Autophagy. Cancers (Basel) 11(10):1422. https://doi.org/10.3390/cancers11101422
Chang YY, Neufeld TP (2009) An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20(7):2004–2014. https://doi.org/10.1091/mbc.e08-12-1250
Manzoni C, Mamais A, Dihanich S et al (2018) mTOR Independent alteration in ULK1 Ser758 phosphorylation following chronic LRRK2 kinase inhibition. Biosci Rep 38(2):BSR20171669. https://doi.org/10.1042/BSR20171669
Tamargo-Gómez I, Mariño G (2018) AMPK: Regulation of Metabolic dynamics in the Context of Autophagy. Int J Mol Sci 19(12):3812. https://doi.org/10.3390/ijms19123812
Backer JM (2016) The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J 473(15):2251–2271. https://doi.org/10.1042/BCJ20160170
Altomare DA, Wang HQ, Skele KL et al (2004) AKT and mTOR phosphorylation is frequently detected in Ovarian cancer and can be targeted to disrupt ovarian Tumor cell growth. Oncogene 23(34):5853–5857. https://doi.org/10.1038/sj.onc.1207721
Kudo Y, Sugimoto M, Arias E et al (2020) PKCλ/ι loss induces Autophagy, oxidative phosphorylation, and NRF2 to promote Liver Cancer Progression. Cancer Cell 38(2):247–262e11. https://doi.org/10.1016/j.ccell.2020.05.018
Klionsky DJ, Schulman BA (2014) Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 21(4):336–345. https://doi.org/10.1038/nsmb.2787
Tracz M, Bialek W (2021) Beyond K48 and K63: non-canonical protein ubiquitination. Cell Mol Biol Lett 26(1):1. https://doi.org/10.1186/s11658-020-00245-6
Manley S, Williams JA, Ding WX (2013) Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med (Maywood) 238(5):525–538. https://doi.org/10.1177/1535370213489446
Shin WH, Park JH, Chung KC (2020) The central regulator p62 between ubiquitin proteasome system and autophagy and its role in the mitophagy and Parkinson’s Disease. BMB Rep 53(1):56–63. https://doi.org/10.5483/BMBRep.2020.53.1.283
Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C (2014) P62/SQSTM1 at the interface of aging, autophagy, and Disease. Age (Dordr) 36(3):9626. https://doi.org/10.1007/s11357-014-9626-3
Rider L, Cramer SD (2015) SPOP the mutation. Elife 4:e11760. https://doi.org/10.7554/eLife.11760
Gao K, Shi Q, Liu Y, Wang C (2022) Enhanced autophagy and NFE2L2/NRF2 pathway activation in SPOP mutation-driven Prostate cancer. Autophagy 18(8):2013–2015. https://doi.org/10.1080/15548627.2022.2062873
Raimondi M, Cesselli D, Di Loreto C, La Marra F, Schneider C, Demarchi F (2019) USP1 (ubiquitin specific peptidase 1) targets ULK1 and regulates its cellular compartmentalization and autophagy. Autophagy 15(4):613–630. https://doi.org/10.1080/15548627.2018.1535291
Kim JH, Seo D, Kim SJ et al (2018) The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep 19(4):e44378. https://doi.org/10.15252/embr.201744378
Han SH, Korm S, Han YG et al (2019) GCA links TRAF6-ULK1-dependent autophagy activation in resistant chronic Myeloid Leukemia. Autophagy 15(12):2076–2090. https://doi.org/10.1080/15548627.2019.1596492
Cao L, Liu X, Zheng B, Xing C, Liu J (2022) Role of K63-linked ubiquitination in cancer. Cell Death Discov 8(1):410. https://doi.org/10.1038/s41420-022-01204-0
Han T, Guo M, Gan M, Yu B, Tian X, Wang JB (2018) TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy 14(12):2035–2048. https://doi.org/10.1080/15548627.2018.1491493
Zhan Z, Xie X, Cao H et al (2014) Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of Lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy 10(2):257–268. https://doi.org/10.4161/auto.27162
Wang G, Gao Y, Li L et al (2012) K63-linked ubiquitination in kinase activation and cancer. Front Oncol 2:5. https://doi.org/10.3389/fonc.2012.00005
Chen YH, Huang TY, Lin YT et al (2021) VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. Nat Commun 12(1):1322. https://doi.org/10.1038/s41467-021-21715-1
Liu Z, Chen P, Gao H et al (2014) Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for Tumor suppression. Cancer Cell 26(1):106–120. https://doi.org/10.1016/j.ccr.2014.05.015
Xia Q, Ali S, Liu L et al (2020) Role of Ubiquitination in PTEN Cellular Homeostasis and its implications in GB Drug Resistance. Front Oncol 10:1569. https://doi.org/10.3389/fonc.2020.01569
Zhai F, Wang J, Yang W, Ye M, Jin X (2022) The E3 ligases in Cervical Cancer and Endometrial Cancer. Cancers (Basel) 14(21):5354. https://doi.org/10.3390/cancers14215354
Lee YR, Chen M, Lee JD et al (2019) Reactivation of PTEN Tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 364(6441):eaau0159. https://doi.org/10.1126/science.aau0159
Boughton AJ, Krueger S, Fushman D (2020) Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and enhanced Affinity for Proteasomal Subunit Rpn1. Structure 28(1):29–43e6. https://doi.org/10.1016/j.str.2019.10.008
Lee MS, Jeong MH, Lee HW et al (2015) PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun 6:7769. https://doi.org/10.1038/ncomms8769
Xu Y, Wan W (2023) Acetylation in the regulation of autophagy. Autophagy 19(2):379–387. https://doi.org/10.1080/15548627.2022.2062112
Lee IH (2019) Mechanisms and Disease implications of sirtuin-mediated autophagic regulation. Exp Mol Med 51(9):1–11. https://doi.org/10.1038/s12276-019-0302-7
Lee D, Goldberg AL (2013) SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J Biol Chem 288(42):30515–30526. https://doi.org/10.1074/jbc.M113.489716
Kim JY, Mondaca-Ruff D, Singh S, Wang Y (2022) SIRT1 and autophagy: implications in Endocrine Disorders. Front Endocrinol (Lausanne) 13:930919. https://doi.org/10.3389/fendo.2022.930919
Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115(6):727–738. https://doi.org/10.1016/s0092-8674(03)00939-5
Zhang X, Yuan Z, Zhang Y et al (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell 27(2):197–213. https://doi.org/10.1016/j.molcel.2007.05.033
Cao Y, Luo Y, Zou J et al (2019) Autophagy and its role in gastric cancer. Clin Chim Acta 489:10–20. https://doi.org/10.1016/j.cca.2018.11.028
Sun J, Tai S, Tang L et al (2021) Acetylation Modification during Autophagy and Vascular Aging. Front Physiol 12:598267. https://doi.org/10.3389/fphys.2021.598267
Lin SY, Li TY, Liu Q et al (2012) GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336(6080):477–481. https://doi.org/10.1126/science.1217032
Nie T, Yang S, Ma H et al (2016) Regulation of ER stress-induced autophagy by GSK3β-TIP60-ULK1 pathway. Cell Death Dis 7(12):e2563. https://doi.org/10.1038/cddis.2016.423
Su H, Yang F, Wang Q et al (2017) VPS34 acetylation controls its lipid kinase activity and the initiation of Canonical and non-canonical autophagy. Mol Cell 67(6):907–921e7. https://doi.org/10.1016/j.molcel.2017.07.024
Rao Y, Wan Q, Su H et al (2018) ROS-induced HSP70 promotes cytoplasmic translocation of high-mobility group box 1b and stimulates antiviral autophagy in grass carp kidney cells. J Biol Chem 293(45):17387–17401. https://doi.org/10.1074/jbc.RA118.003840
Marquez RT, Xu L (2012) Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res 2(2):214–221 PMID: 22485198
Sun T, Ming L, Yan Y, Zhang Y, Xue H (2017) Beclin 1 acetylation impairs the anticancer effect of aspirin in Colorectal cancer cells. Oncotarget 8(43):74781–74790. https://doi.org/10.18632/oncotarget.20367
Huang R, Xu Y, Wan W et al (2015) Deacetylation of nuclear LC3 drives autophagy initiation under Starvation. Mol Cell 57(3):456–466. https://doi.org/10.1016/j.molcel.2014.12.013
Liu J, Bi X, Chen T et al (2015) Shear stress regulates endothelial cell autophagy via redox regulation and Sirt1 expression. Cell Death Dis 6(7):e1827. https://doi.org/10.1038/cddis.2015.193
Yu Z, Li H, Zhu J, Wang H, Jin X (2022) The roles of E3 ligases in hepatocellular carcinoma. Am J Cancer Res 12(3):1179–1214 PMID: 35411231
Wang T, Zhang X, Li JJ (2002) The role of NF-kappaB in the regulation of cell stress responses. Int Immunopharmacol 2(11):1509–1520. https://doi.org/10.1016/s1567-5769(02)00058-9
Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B et al (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11(5):407–420. https://doi.org/10.1016/j.ccr.2007.04.001
Zeng Z, Liang J, Wu L, Zhang H, Lv J, Chen N (2020) Exercise-Induced Autophagy suppresses Sarcopenia through Akt/mTOR and Akt/FoxO3a Signal Pathways and AMPK-Mediated mitochondrial Quality Control. Front Physiol 11:583478. https://doi.org/10.3389/fphys.2020.583478
Yu X, Ma R, Wu Y, Zhai Y, Li S (2018) Reciprocal regulation of metabolic reprogramming and epigenetic modifications in Cancer. Front Genet 9:394. https://doi.org/10.3389/fgene.2018.00394
Wan L, Xu K, Wei Y et al (2018) Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function. Mol Cell 69(2):279–291e5. https://doi.org/10.1016/j.molcel.2017.12.024
Chang C, Su H, Zhang D et al (2015) AMPK-Dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for Autophagy upon glucose Starvation. Mol Cell 60(6):930–940. https://doi.org/10.1016/j.molcel.2015.10.037
Shi Y, Shen HM, Gopalakrishnan V, Gordon N (2021) Epigenetic regulation of Autophagy beyond the cytoplasm: a review. Front Cell Dev Biol 9:675599. https://doi.org/10.3389/fcell.2021.675599
Sakamaki JI, Wilkinson S, Hahn M et al (2017) Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol Cell 66(4):517–532e9. https://doi.org/10.1016/j.molcel.2017.04.027
Wang Y, Huang Y, Liu J et al (2020) Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep 21(1):e48335. https://doi.org/10.15252/embr.201948335
Lu L, Li L, Lv X, Wu XS, Liu DP, Liang CC (2011) Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res 21(8):1182–1195. https://doi.org/10.1038/cr.2011.71
Wei FZ, Cao Z, Wang X et al (2015) Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy 11(12):2309–2322. https://doi.org/10.1080/15548627.2015.1117734
Popova EY, Pinzon-Guzman C, Salzberg AC et al (2016) LSD1-Mediated demethylation of H3K4me2 is required for the transition from late progenitor to differentiated mouse Rod Photoreceptor. Mol Neurobiol 53(7):4563–4581. https://doi.org/10.1007/s12035-015-9395-8
Li L, Liu W, Sun Q, Zhu H, Hong M, Qian S (2021) Decitabine Downregulates TIGAR to Induce Apoptosis and Autophagy in Myeloid Leukemia Cells. Oxid Med Cell Longev. 2021:8877460. https://doi.org/10.1155/2021/8877460
Schnekenburger M, Grandjenette C, Ghelfi J et al (2011) Sustained exposure to the DNA demethylating agent, 2’-deoxy-5-azacytidine, leads to apoptotic cell death in chronic Myeloid Leukemia by promoting differentiation, senescence, and autophagy. Biochem Pharmacol 81(3):364–378. https://doi.org/10.1016/j.bcp.2010.10.013
Yang PM, Lin YT, Shun CT et al (2013) Zebularine inhibits tumorigenesis and stemness of Colorectal cancer via p53-dependent endoplasmic reticulum stress. Sci Rep 3:3219. https://doi.org/10.1038/srep03219
Gao L, Sun X, Zhang Q et al (2018) Histone deacetylase inhibitor trichostatin A and autophagy inhibitor chloroquine synergistically exert anti-tumor activity in H-ras transformed breast epithelial cells. Mol Med Rep 17(3):4345–4350. https://doi.org/10.3892/mmr.2018.8446
Bai Y, Chen Y, Chen X et al (2019) Trichostatin A activates FOXO1 and induces autophagy in osteosarcoma. Arch Med Sci 15(1):204–213. https://doi.org/10.5114/aoms.2018.73860
Lee JY, Kuo CW, Tsai SL et al (2016) Inhibition of HDAC3- and HDAC6-Promoted Survivin expression plays an important role in SAHA-Induced autophagy and viability reduction in Breast Cancer cells. Front Pharmacol 7:81. https://doi.org/10.3389/fphar.2016.00081
Sun J, Piao J, Li N, Yang Y, Kim KY, Lin Z (2020) Valproic acid targets HDAC1/2 and HDAC1/PTEN/Akt signalling to inhibit cell proliferation via the induction of autophagy in gastric cancer. FEBS J 287(10):2118–2133. https://doi.org/10.1111/febs.15122
Dong LH, Cheng S, Zheng Z et al (2013) Histone deacetylase inhibitor potentiated the ability of MTOR inhibitor to induce autophagic cell death in Burkitt leukemia/lymphoma. J Hematol Oncol 6:53. https://doi.org/10.1186/1756-8722-6-53
Yang F, Wang F, Liu Y et al (2018) Sulforaphane induces autophagy by inhibition of HDAC6-mediated PTEN activation in triple negative Breast cancer cells. Life Sci 213:149–157. https://doi.org/10.1016/j.lfs.2018.10.034
De U, Son JY, Sachan R et al (2018) A New Synthetic histone deacetylase inhibitor, MHY2256, induces apoptosis and Autophagy Cell Death in Endometrial Cancer cells via p53 acetylation. Int J Mol Sci 19(9):2743. https://doi.org/10.3390/ijms19092743
Xu H, Zhang L, Qian X et al (2019) GSK343 induces autophagy and downregulates the AKT/mTOR signaling pathway in Pancreatic cancer cells. Exp Ther Med 18(4):2608–2616. https://doi.org/10.3892/etm.2019.7845
Li R, Yi X, Wei X et al (2018) EZH2 inhibits autophagic cell death of aortic vascular smooth muscle cells to affect Aortic Dissection. Cell Death Dis 9(2):180. https://doi.org/10.1038/s41419-017-0213-2
Haebe JR, Bergin CJ, Sandouka T, Benoit YD (2021) Emerging role of G9a in cancer stemness and promises as a therapeutic target. Oncogenesis 10(11):76. https://doi.org/10.1038/s41389-021-00370-7
Kim TW, Cheon C, Ko SG (2020) SH003 activates autophagic cell death by activating ATF4 and inhibiting G9a under hypoxia in gastric cancer cells. Cell Death Dis 11(8):717. https://doi.org/10.1038/s41419-020-02924-w
Kim TW, Lee SY, Kim M, Cheon C, Ko SG (2018) Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis 9(9):875. https://doi.org/10.1038/s41419-018-0930-1
Chao A, Lin CY, Chao AN et al (2017) Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies. Oncotarget 8(43):74434–74450. https://doi.org/10.18632/oncotarget.20158
Wu K, Woo SM, Kwon TK (2020) The histone lysine-specific demethylase 1 inhibitor, SP2509 exerts cytotoxic effects against Renal Cancer cells through downregulation of Bcl-2 and Mcl-1. J Cancer Prev 25(2):79–86. https://doi.org/10.15430/JCP.2020.25.2.79
Wen X, Klionsky DJ (2017) BRD4 is a newly characterized transcriptional regulator that represses autophagy and lysosomal function. Autophagy 13(11):1801–1803. https://doi.org/10.1080/15548627.2017.1364334
Jang JE, Eom JI, Jeung HK et al (2017) AMPK-ULK1-Mediated Autophagy confers resistance to BET inhibitor JQ1 in Acute Myeloid Leukemia stem cells. Clin Cancer Res 23(11):2781–2794. https://doi.org/10.1158/1078-0432.CCR-16-1903
Li W, Shen X, Feng S, Liu Y, Zhao H et al (2022) BRD4 inhibition by JQ1 protects against LPS-induced cardiac dysfunction by inhibiting activation of NLRP3 inflammasomes. Mol Biol Rep 49(9):8197–8207. https://doi.org/10.1007/s11033-022-07377-2
Luan W, Pang Y, Li R et al (2019) Akt/mTOR-Mediated Autophagy confers Resistance to BET inhibitor JQ1 in Ovarian Cancer. Onco Targets Ther 12:8063–8074. https://doi.org/10.2147/OTT.S220267
Acknowledgements
The authors would like to acknowledge the academic fellowship grant from the Department of Science and Technology and Biotechnology, Government of West Bengal.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors reviewed the manuscript. The authors declare that all data were generated in-house and that no paper mill was used.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
All the authors approved the publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chowdhury, S.G., Karmakar, P. Revealing the role of epigenetic and post-translational modulations of autophagy proteins in the regulation of autophagy and cancer: a therapeutic approach. Mol Biol Rep 51, 3 (2024). https://doi.org/10.1007/s11033-023-08961-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-08961-w