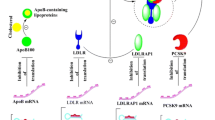
Abstract
Background
Familial hypercholesterolemia (FH) is caused by pathogenic variants in low-density lipoprotein (LDL) receptor (LDLR) or its associated genes, including apolipoprotein B (APOB), proprotein convertase subtilisin/kexin type 9 (PCSK9), and LDLR adaptor protein 1 (LDLRAP1). However, approximately 40% of the FH patients clinically diagnosed (based on FH phenotypes) may not carry a causal variant in a FH-related gene. Variants located at 3’ untranslated region (UTR) of FH-related genes could elucidate mechanisms involved in FH pathogenesis. This study used a computational approach to assess the effects of 3’UTR variants in FH-related genes on miRNAs molecular interactions and to explore the association of these variants with molecular diagnosis of FH.
Methods and results
Exons and regulatory regions of FH-related genes were sequenced in 83 FH patients using an exon-target gene sequencing strategy. In silico prediction tools were used to study the effects of 3´UTR variants on interactions between miRNAs and target mRNAs. Pathogenic variants in FH-related genes (molecular diagnosis) were detected in 44.6% FH patients. Among 59 3’UTR variants identified, LDLR rs5742911 and PCSK9 rs17111557 were associated with molecular diagnosis of FH, whereas LDLR rs7258146 and rs7254521 and LDLRAP1 rs397860393 had an opposite effect (p < 0.05). 3´UTR variants in LDLR (rs5742911, rs7258146, rs7254521) and PCSK9 (rs17111557) disrupt interactions with several miRNAs, and more stable bindings were found with LDLR (miR-4435, miR-509-3 and miR-502) and PCSK9 (miR-4796).
Conclusion
LDLR and PCSK9 3´UTR variants disturb miRNA:mRNA interactions that could affect gene expression and are potentially associated with molecular diagnosis of FH.


Similar content being viewed by others
Data availability
All relevant data of this study are available within the article and its Supplementary Information. The raw DNA sequence reads (BioProject accession number PRJNA662090) are available for download at https://www.ncbi.nlm.nih.gov/sra/PRJNA662090. Source data are provided with this paper.
References
Akioyamen LE, Genest J, Chu A et al (2019) Risk factors for cardiovascular disease in heterozygous familial hypercholesterolemia: a systematic review and meta-analysis. J Clin Lipidol 13:15–30. https://doi.org/10.1016/j.jacl.2018.10.012
Abifadel M, Boileau C (2023) Genetic and molecular architecture of familial hypercholesterolemia. J Intern Med 293:144–165
Chen P, Chen X, Zhang S (2019) Current status of familial hypercholesterolemia in China: a need for patient FH registry systems. Front Physiol 10. https://doi.org/10.3389/fphys.2019.00280
Sturm AC, Knowles JW, Gidding SS et al (2018) Clinical genetic testing for familial hypercholesterolemia. J Am Coll Cardiol 72:662–680. https://doi.org/10.1016/j.jacc.2018.05.044
Berberich AJ, Hegele RA (2019) The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol 16:9–20. https://doi.org/10.1038/s41569-018-0052-6
Nordestgaard BG, Chapman MJ, Humphries SE et al (2013) Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the european atherosclerosis society. Eur Heart J 34:3478–90a. https://doi.org/10.1093/eurheartj/eht273
Pirazzi C, Håkansson L, Gustafsson C et al (2019) < p > high prevalence of genetic determined familial hypercholesterolemia in premature coronary artery disease. Appl Clin Genet Volume 12:71–78. https://doi.org/10.2147/TACG.S202942
Chora JR, Medeiros AM, Alves AC, Bourbon M (2018) Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet Sci 20:591–598. https://doi.org/10.1038/gim.2017.151
Steri M, Idda ML, Whalen MB, Orrù V (2018) Genetic variants in mRNA untranslated regions. Wiley Interdiscip Rev RNA 9:1–20. https://doi.org/10.1002/wrna.1474
Heshmatzad K, Naderi N, Maleki M et al (2023) Role of non-coding variants in cardiovascular disease. J Cell Mol Med 27:1621–1636
Rojano E, Seoane P, Ranea JAG, Perkins JR (2019) Regulatory variants: from detection to predicting impact. Brief Bioinform 20:1639–1654. https://doi.org/10.1093/BIB/BBY039
de Araújo JNG, de Oliveira VF, Borges JB et al (2022) In silico analysis of upstream variants in brazilian patients with familial hypercholesterolemia. Gene 849:146908. https://doi.org/10.1016/j.gene.2022.146908
Zhou S, Jin J, Wang J et al (2018) miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 39:1073–1084. https://doi.org/10.1038/aps.2018.30
Moszyńska A, Gebert M, Collawn JF, Bartoszewski R (2017) SNPs in microRNA target sites and their potential role in human disease. Open Biol 7. https://doi.org/10.1098/rsob.170019
Bruno AE, Li L, Kalabus JL et al (2012) miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3’UTRs of human genes. BMC Genomics 13:44. https://doi.org/10.1186/1471-2164-13-44
Van Zyl T, Jerling JC, Conradie KR, Feskens EJ (2014) Common and rare single nucleotide polymorphisms in the LDLR gene are present in a black south african population and associate with low-density lipoprotein cholesterol levels. J Hum Genet 59:88–94. https://doi.org/10.1038/jhg.2013.123
Zambrano T, Hirata MH, Cerda Á et al (2015) Impact of 3’UTR genetic variants in PCSK9 and LDLR genes on plasma lipid traits and response to atorvastatin in brazilian subjects: a pilot study. Int J Clin Exp Med 8:5978–5988
Los B, Borges JB, Oliveira VF et al (2021) Functional analysis of PCSK9 3′UTR variants and mRNA–miRNA interactions in patients with familial hypercholesterolemia. Epigenomics 13:779–791. https://doi.org/10.2217/epi-2020-0462
Decourt C, Janin A, Moindrot M et al (2020) PCSK9 post-transcriptional regulation: role of a 3′UTR microRNA-binding site variant in linkage disequilibrium with c.1420G. Atherosclerosis 314:63–70. https://doi.org/10.1016/j.atherosclerosis.2020.10.010
Pérez-Campo FM, De Castro-Orós I, Noriega A et al (2017) Functional analysis of new 3′ untranslated regions genetic variants in genes associated with genetic hypercholesterolemias. J Clin Lipidol 11:532–542. https://doi.org/10.1016/j.jacl.2017.02.004
Faludi A, Izar M, Saraiva J, DA DIRETRIZ BRASILEIRA DE DISLIPIDEMIAS E PREVENÇÃO DA ATEROSCLEROSE – 2017 (2017) ATUALIZAÇÃO. Arq Bras Cardiol 109:. https://doi.org/10.5935/abc.20170121
World Health Organization. WHO (1998) Human Genetics Programme. Familial Hypercholesterolaemia (FH). Report of a second WHO Consultation. Geneva
Borges JB, de Oliveira VF, Ferreira GM et al (2021) Genomics, epigenomics and pharmacogenomics of familial hypercholesterolemia (FHBGEP): a study protocol. Res Social Administrative Pharm 17:1347–1355. https://doi.org/10.1016/j.sapharm.2020.10.007
Borges JB, Oliveira VF, Dagli-Hernandez C et al (2023) Identification of pathogenic variants in the brazilian cohort with familial hypercholesterolemia using exon-targeted gene sequencing. Gene 875:147501. https://doi.org/10.1016/j.gene.2023.147501
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Sci 17:405–424. https://doi.org/10.1038/gim.2015.30
Liu C, Zhang F, Li T et al (2012) MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics 13. https://doi.org/10.1186/1471-2164-13-661
Barenboim M, Zoltick BJ, Guo Y, Weinberger DR (2010) MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat 31:1223–1232. https://doi.org/10.1002/humu.21349
Sun H, Nicoloso MS, Bhattacharyya A et al (2008) In silico prediction of target SNPs affecting miR-mRNA interaction. In: 2008 IEEE International Conference on Bioinformatics and Biomeidcine Workshops. IEEE, pp 211–214
Bhattacharya A, Ziebarth JD, Cui Y (2014) PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 42:86–91. https://doi.org/10.1093/nar/gkt1028
Moldovan V, Banescu C, Dobreanu M (2020) Molecular diagnosis methods in familial hypercholesterolemia. Anatol J Cardiol 23:120–127. https://doi.org/10.14744/ANATOLJCARDIOL.2019.95038
Sun H, Yu G (2019) New insights into the pathogenicity of non-synonymous variants through multi-level analysis. Sci Rep 9:1667. https://doi.org/10.1038/s41598-018-38189-9
Lorenzo A, da De, Silva JDL, James CE et al (2017) Clinical, anthropometric and biochemical characteristics of patients with or without genetically confirmed familial hypercholesterolemia. Arq Bras Cardiol 110:119–123. https://doi.org/10.5935/abc.20180005
Rodríguez-Arroyo G, Paradisi I, Vívenes-Lugo M et al (2016) Polimorfismos de los genes LEP, LDLR, APOA4 y sus relaciones con el sobrepeso, la obesidad y el riesgo de enfermedades crónicas en adultos del estado Sucre. Venezuela Biomed 36:78–90. https://doi.org/10.7705/biomedica.v36i1.2702
Orang AV, Safaralizadeh R, Kazemzadeh-Bavili M (2014) Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014:. https://doi.org/10.1155/2014/970607
Lewis BP, Shih I, Jones-Rhoades MW et al (2003) Prediction of mammalian microRNA targets. Cell 115:787–798. https://doi.org/10.1016/S0092-8674(03)01018-3
Xue Y, Yin P, Li G, Zhong D (2018) Genome-wide integration study of circulating miRNAs and peripheral whole-blood mRNAs of male Acute Ischemic Stroke Patients. Neuroscience 380:27–37. https://doi.org/10.1016/j.neuroscience.2018.04.001
Raitoharju E, Seppälä I, Oksala N et al (2014) Blood microRNA profile associates with the levels of serum lipids and metabolites associated with glucose metabolism and insulin resistance and pinpoints pathways underlying metabolic syndrome. The cardiovascular risk in Young Finns Study. Mol Cell Endocrinol 391:41–49. https://doi.org/10.1016/j.mce.2014.04.013
Mangravite LM, Medina MW, Cui J et al (2010) Combined influence of LDLR and HMGCR sequence variation on lipid-lowering response to simvastatin. Arterioscler Thromb Vasc Biol 30:1485–1492. https://doi.org/10.1161/ATVBAHA.110.203273
Carr G, Barrese V, Stott JB et al (2016) MicroRNA-153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension. Cardiovasc Res 112:581–589. https://doi.org/10.1093/cvr/cvw177
Zou Y, Liu W, Zhang J, Xiang D (2016) MiR-153 regulates apoptosis and autophagy of cardiomyocytes by targeting Mcl-1. Mol Med Rep 14:1033–1039. https://doi.org/10.3892/mmr.2016.5309
Lu Y, Li Y, Li G, Lu H (2020) Identification of potential markers for type 2 diabetes mellitus via bioinformatics analysis. Mol Med Rep 22:1868–1882. https://doi.org/10.3892/mmr.2020.11281
Paquette M, Bernard S, Ruel I et al (2019) Diabetes is associated with an increased risk of cardiovascular disease in patients with familial hypercholesterolemia. J Clin Lipidol 13:123–128. https://doi.org/10.1016/J.JACL.2018.09.008
Sepramaniam S, Tan JR, Tan KS et al (2014) Circulating MicroRNAs as biomarkers of acute stroke. Int J Mol Sci 15:1418–1432. https://doi.org/10.3390/ijms15011418
Janaszak-Jasiecka A, Siekierzycka A, Bartoszewska S et al (2018) eNOS expression and NO release during hypoxia is inhibited by miR-200b in human endothelial cells. Angiogenesis 21:711–724. https://doi.org/10.1007/s10456-018-9620-y
Xu S, Ilyas I, Little PJ et al (2021) Endothelial dysfunction in atherosclerotic Cardiovascular Diseases and Beyond: from mechanism to Pharmacotherapies. Pharmacol Rev 73:924–967. https://doi.org/10.1124/PHARMREV.120.000096
Sun Y, Zhang X, Gao H et al (2019) Expression of microRNA-514a-5p and its biological function in experimental pulmonary thromboembolism. Am J Transl Res 11:5514–5530
Kuniholm MH, Liang H, Anastos K et al (2017) Association of a 3′ untranslated region polymorphism in proprotein convertase subtilisin/kexin type 9 with HIV viral load and CD4 + levels in HIV/hepatitis C virus coinfected women. AIDS 31:2483–2492. https://doi.org/10.1097/QAD.0000000000001648
Rosenson RS, Hegele RA, Koenig W (2019) Cholesterol-lowering agents PCSK9 inhibitors today and tomorrow. Circ Res 124:364–385
Gibson G (2012) Rare and common variants: twenty arguments. Nature Reviews Genetics 2012 13:2 13:135–145. https://doi.org/10.1038/nrg3118
Acknowledgements
The authors thank Adriana Garofalo, Cristina Moreno Fajardo and other colleagues at Laboratory of Molecular Research in Cardiology and Dyslipidemia Division of the Institute of Cardiology Dante Pazzanese for technical support and assistance in patient selection and data collection. RCCF, RHB, CDH and VFO, are grateful to Sao Paulo Research Foundation (FAPESP) Research Fellowship Program, Brazil. ESRM, ADL, RDCH and MHH are grateful to National Council for Scientific and Technological Development (CNPq) Research Fellowship Program, Brazil.
Funding
This study is funded by Sao Paulo Research Foundation (FAPESP, # 2016/12899-6), National Council for Scientific and Technological Development (CNPq, # 447120/2014-0), and Coordination of Superior Level Staff Improvement (CAPES, # code001), Brazil.
Author information
Authors and Affiliations
Contributions
MHH, RCCF, RHB and RDCH conceived and designed the study. RMG and AAF conducted the selection, clinical evaluation and follow-up of FH patients. RCCF, CDH, ESRM, GMB and VNS acquired and analyzed clinical data. JBB and VFO performed DNA sequencing and bioinformatics data analysis. RCCF performed in silico studies and data analysis and interpretation. RCCF, RHB and ADL performed the statistical analysis. RCCF and RHB drafted the manuscript. MHH, RDCH, ADL and VNS critically reviewed the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial and non-financial interests to disclose.
Ethics approval
The study protocol was approved by Local Ethics Committees (CAAE 24618713.0.0000.5462, 24618713.0.1001.5462, 24618713.0.3001.0067 and 24618713.0.2001.5292) and conducted according to good clinical practices and the Declaration of Helsinki guidelines (as revised in 2013).
Consent to participate
Informed consent was collected from all subjects who participated in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Freitas, R.C.C., Bortolin, R.H., Borges, J.B. et al. LDLR and PCSK9 3´UTR variants and their putative effects on microRNA molecular interactions in familial hypercholesterolemia: a computational approach. Mol Biol Rep 50, 9165–9177 (2023). https://doi.org/10.1007/s11033-023-08784-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08784-9