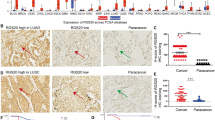
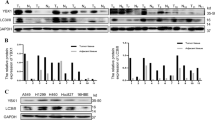
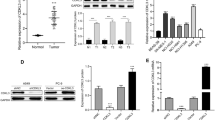
Abstract
Background
Non-small cell lung carcinoma (NSCLC) is the most common cause of cancer-associated deaths worldwide. Though recent development in targeted therapy has improved NSCLC prognosis, yet there is an unmet need to identify novel causative factors and appropriate therapeutic regimen against NSCLCs.
Methods and results
In this study, we identify key molecular factors de-regulated in NSCLCs. Analyze their expression by real-time PCR and immunoblot; map their localization by immuno-fluorescence microscopy. We further propose an FDA approved drug, chloroquine (CQ) that affects the function of the molecular factors and hence can be repurposed as a therapeutic strategy against NSCLCs. Available NSCLC mutation data reflects a high probabilistic chance of patients harboring a p53 mutation, especially a gain of function (GOF)-R273H mutation. The GOF-P53 mutation enables the P53 protein to potentially interact with non-canonical protein partners facilitating oncogenesis. In this context, analysis of existing transcriptomic data from R273H-P53 expressing cells shows a concomitant up-regulation of Yes-associated protein (YAP) transcriptional targets and its protein partner TEAD1 in NSCLCs, suggesting a possible link between R273H-P53 and YAP. We therefore explored the inter-dependence of R273H-P53 and YAP in NSCLC cells. They were found to co-operatively regulate NSCLC proliferation. Genetic or pharmacological inhibition of YAP and GOF-P53 resulted in sensitization of NSCLC cells. Further analysis of pathways controlled by GOF-P53 and YAP showed that they positively regulate the cellular homeostatic process- autophagy to mediate survival. We hence postulated that a modulation of autophagy might be a potent strategy to curb proliferation. In accordance to above, autophagy inhibition, especially with the FDA-approved drug- chloroquine (CQ) resulted in cytoplasmic accumulation and reduced transcriptional activity of GOF-P53 and YAP, leading to growth arrest of NSCLC cells.
Conclusion
Our study highlights the importance of GOF-P53 and YAP in NSCLC proliferation and proposes autophagy inhibition as an efficient strategy to attenuate NSCLC tumorigenesis.





Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BAF:
-
Bafilomycin
- CMA:
-
Chaperone mediated autophagy
- CQ:
-
Chloroquine
- EGFR:
-
Epidermal growth factor receptor
- EV:
-
Empty vector
- GDC:
-
Genomic data commons
- GOF:
-
Gain of function
- LTR:
-
Lysotracker red
- MDC:
-
Monodansylcadaverine
- NSCLC:
-
Non-small cell lung carcinoma
- PFT-α:
-
Pifithrin-alpha
- SCLC:
-
Small-cell lung carcinoma
- VP:
-
Verteporfin
- WT:
-
Wild type
- YAP:
-
Yes-associated protein
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2020) Global cancer statistics: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. https://doi.org/10.3322/caac.21660
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H et al (2010) Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med 362(25):2380–2388. https://doi.org/10.1056/NEJMoa0909530
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J et al (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11(2):121–128. https://doi.org/10.1016/S1470-2045(09)70364-X
Yoh K, Seto T, Satouchi M, Nishio M, Yamamoto N, Murakami H et al (2017) Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 5(1):42–50
Shaw AT, Ou S-HI, Bang Y-J, Camidge DR, Solomon BJ, Salgia R et al (2014) Crizotinib in ROS1-rearranged non–small-cell lung cancer. N Engl J Med 371(21):1963–1971. https://doi.org/10.1056/NEJMoa1406766
Shaw AT, Kim D-W, Nakagawa K, Seto T, Crinó L, Ahn M-J et al (2013) Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 368(25):2385–2394. https://doi.org/10.1056/NEJMoa1214886
Socinski MA, Evans T, Gettinger S, Hensing TA, Sequist LV, Ireland B et al (2013) Treatment of stage IV non-small cell lung cancer: diagnosis and management of lung cancer: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143(5):e341S–e68. https://doi.org/10.1378/chest.12-2361
Liu W-j, Du Y, Wen R, Yang M, therapeutics Xu JJP (2020) Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Cell Death Dis 206:107438. https://doi.org/10.1016/j.pharmthera.2019.107438
Petitjean A, Achatz M, Borresen-Dale A, Hainaut P, Olivier M (2007) TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 26(15):2157–2165. https://doi.org/10.1038/sj.onc.1210302
Harris CC, Hollstein M (1993) Clinical implications of the p53 tumor-suppressor gene. N Engl J Med 329(18):1318–1327. https://doi.org/10.1056/NEJM199310283291807
Kim MP, Lozano GJCD, Differentiation (2018) Mutant p53 partners in crime. Cell Death Differ 25(1):161–168. https://doi.org/10.1038/cdd.2017.185
Soussi T, Lozano G (2005) p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun 331(3):834–842. https://doi.org/10.1016/j.bbrc.2005.03.190
Vaughan CA, Singh S, Grossman SR, Windle B, Deb SP, Deb SJMo (2017) Gain-of‐function p53 activates multiple signaling pathways to induce oncogenicity in lung cancer cells. Mol Oncol 11(6):696–711. https://doi.org/10.1002/1878-0261.12068
Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino GJO (2007) Mutant p53: an oncogenic transcription factor. Oncogene 26(15):9. https://doi.org/10.1038/sj.onc.1210296
Rasheduzzaman M, Jeong J-K, Park S-YJLs (2018) Resveratrol sensitizes lung cancer cell to TRAIL by p53 independent and suppression of Akt/NF-κB signaling. Life Sci 208:208–220. https://doi.org/10.1016/j.lfs.2018.07.035
Yang X, Zhang Q, Yang X, Zhao M, Yang T, Yao A et al (2019) PACT cessation overcomes ovarian cancer cell chemoresistance to cisplatin by enhancing p53-mediated apoptotic pathway. Biochem Biophys Res Commun 511(4):719–724. https://doi.org/10.1016/j.bbrc.2019.02.089
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. AACR. https://doi.org/10.1158/2159-8290.CD-12-0095
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):pl1–pl. https://doi.org/10.1126/scisignal.2004088
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G et al (2017) A pathology atlas of the human cancer transcriptome. Science. https://doi.org/10.1126/science.aan2507
Saini H, Hakeem I, Mukherjee S, Chowdhury S, Chowdhury R (2019) Autophagy regulated by gain of function mutant p53 enhances proteasomal inhibitor-mediated cell death through induction of ROS and ERK in lung cancer cells. J Oncol. https://doi.org/10.1155/2019/6164807
Saini H, Sharma H, Mukherjee S, Chowdhury S, Chowdhury RJCCI (2021) Verteporfin disrupts multiple steps of autophagy and regulates p53 to sensitize osteosarcoma cells. Cancer Cell Int 21(1):1–16. https://doi.org/10.1186/s12935-020-01720-y
Villalta JI, Galli S, Iacaruso MF, Arciuch VGA, Poderoso JJ, Jares-Erijman EA et al (2011) New algorithm to determine true colocalization in combination with image restoration and time-lapse confocal microscopy to MAP kinases in mitochondria. PLoS ONE 6(4):e19031. https://doi.org/10.1371/journal.pone.0019031
Chowdhury R, Chowdhury S, Roychoudhury P, Mandal C, Chaudhuri K (2009) Arsenic induced apoptosis in malignant melanoma cells is enhanced by menadione through ROS generation, p38 signaling and p53 activation. Apoptosis 14(1):108–123. https://doi.org/10.1007/s10495-008-0284-8
Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S et al (2016) YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep 17(2):188–201. https://doi.org/10.15252/embr.201540488
Zucchini C, Manara MC, Cristalli C, Carrabotta M, Greco S, Pinca RS et al (2019) ROCK2 deprivation leads to the inhibition of tumor growth and metastatic potential in osteosarcoma cells through the modulation of YAP activity. J Exp Clin Cancer Res 38(1):1–14. https://doi.org/10.1186/s13046-019-1506-3
Bassi L, Carloni M, Fonti E, De La Peña NP, Meschini R, Palitti FJMRF et al (2002) Pifithrin-α, an inhibitor of p53, enhances the genetic instability induced by etoposide (VP16) in human lymphoblastoid cells treated in vitro. Mutat Res Fundam Mol Mech Mutagen 499(2):163–176. https://doi.org/10.1016/s0027-5107(01)00273-1
Kwon Y, Vinayagam A, Sun X, Dephoure N, Gygi SP, Hong P et al (2013) The Hippo signaling pathway interactome. Science 342(6159):737–740. https://doi.org/10.1126/science.1243971
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema K-J et al (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14(8):1435–1455. https://doi.org/10.1080/15548627.2018.1474314
Liang N, Zhang C, Dill P, Panasyuk G, Pion D, Koka V et al (2014) Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complexYAP is required for tumorigenesis of TSC. J Exp Med 211(11):2249–2263. https://doi.org/10.1084/jem.20140341
Cordani M, Butera G, Pacchiana R, Donadelli MJBeBA-RoC (2017) Molecular interplay between mutant p53 proteins and autophagy in cancer cells. Biochim Biophys Acta Rev Cancer 1867(1):19–28. https://doi.org/10.1016/j.bbcan.2016.11.003
Kaushik S, Massey AC, Mizushima N, Cuervo AM (2008) Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 19(5):2179–2192. https://doi.org/10.1091/mbc.e07-11-1155
Tang Y, Wang X-W, Liu Z-H, Sun Y-M, Tang Y-X, Zhou D-H (2017) Chaperone-mediated autophagy substrate proteins in cancer. Oncotarget 8(31):51970. https://doi.org/10.18632/oncotarget.17583
Kirchner P, Bourdenx M, Madrigal-Matute J, Tiano S, Diaz A, Bartholdy BA et al (2019) Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol 17(5):e3000301. https://doi.org/10.1371/journal.pbio.3000301
Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474(7350):179–183. https://doi.org/10.1038/nature10137
Zhang C, Wang F, Xie Z, Chen L, Sinkemani A, Yu H et al (2018) AMOT 130 linking F-actin to YAP is involved in intervertebral disc degeneration. Cell Prolif 51(6):e12492. https://doi.org/10.1111/cpr.12492
Mizushima N (2004) Methods for monitoring autophagy. Int J Biochem Cell Biol 36(12):2491–2502. https://doi.org/10.1016/j.biocel.2004.02.005
Saha T, Guha D, Manna A, Panda AK, Bhat J, Chatterjee S et al (2016) G-actin guides p53 nuclear transport: potential contribution of monomeric actin in altered localization of mutant p53. Sci Rep 6(1):1–14. https://doi.org/10.1038/srep32626
Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P et al (2017) Universal patterns of selection in cancer and somatic tissues. Cell 171(5):1029–1041 e21. https://doi.org/10.1016/j.cell.2017.09.042
Blandino G, Levine AJ, Oren M (1999) Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18(2):477
Pan DJDc( (2010) The hippo signaling pathway in development and cancer. Dev Cell 19(4):491–505. https://doi.org/10.1016/j.devcel.2010.09.011
Zhao B, Ye X, Yu J, Li L, Li W, Li S et al (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22(14):1962–1971. https://doi.org/10.1101/gad.1664408
Gibault F, Bailly F, Corvaisier M, Coevoet M, Huet G, Melnyk P et al (2017) Molecular features of the YAP inhibitor verteporfin: synthesis of hexasubstituted dipyrrins as potential inhibitors of YAP/TAZ, the downstream effectors of the Hippo pathway. ChemMedChem 12(12):954–961. https://doi.org/10.1002/cmdc.201700063
Zhang W, Gao Y, Li F, Tong X, Ren Y, Han X et al (2015) YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res 75(21):4450–4457
Xu L, Zhang T, Huang W, Liu X, Lu J, Gao X et al (2019) YAP mediates the positive regulation of hnRNPK on the lung adenocarcinoma H1299 cell growth. Acta Biochim Biophys Sin 51(7):677–687. https://doi.org/10.1093/abbs/gmz053
Zhou J, Zhang S, Li Z, Chen Z, Xu Y, Ye W et al (2019) Yap-Hippo promotes A549 lung cancer cell death via modulating MIEF1-related mitochondrial stress and activating JNK pathway. Biomed Pharmacother 113:108754. https://doi.org/10.1016/j.biopha.2019.108754
Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu XJCs (2010) Overexpression of yes-associated protein contributes to progression and poor prognosis of non‐small‐cell lung cancer. Cancer Sci 101(5):1279–1285. https://doi.org/10.1111/j.1349-7006.2010.01511.x
Cheng H, Zhang Z, Rodriguez-Barrueco R, Borczuk A, Liu H, Yu J et al (2016) Functional genomics screen identifies YAP1 as a key determinant to enhance treatment sensitivity in lung cancer cells. Oncotarget 7(20):28976. https://doi.org/10.18632/oncotarget.6721
Wang C, Hu Q, Shen H-MJPr, (2016) Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol Res 105:164–175. https://doi.org/10.1016/j.phrs.2016.01.028
Kimura T, Takabatake Y, Takahashi A, Isaka YJCr (2013) Chloroquine in cancer therapy: a double-edged sword of autophagy. Pharmacol Res 73(1):3–7. https://doi.org/10.1158/0008-5472.CAN-12-2464
Hu J, Cao J, Topatana W, Juengpanich S, Li S, Zhang B et al (2021) Targeting mutant p53 for cancer therapy: direct and indirect strategies. J Hematol Oncol 14(1):1–19. https://doi.org/10.1186/s13045-021-01169-0
Ha J-H, Prela O, Carpizo DR, Loh SNJFiMB, (2022) p53 and zinc: a malleable relationship. Front Mol Biosci. https://doi.org/10.3389/fmolb.2022.895887
Walerych D, Lisek K, Del Sal GJFio (2015) Mutant p53: one, no one, and one hundred thousand. Front Oncol 5:289. https://doi.org/10.3389/fonc.2015.00289
Yamamoto S, Iwakuma TJC (2018) Regulators of oncogenic mutant TP53 gain of function. Cancers 11(1):4. https://doi.org/10.3390/cancers11010004
Xu J, Patel NH, Gewirtz DAJIJoMS (2020) Triangular relationship between p53, autophagy, and chemotherapy resistance. Int J Mol Sci 21(23):8991. https://doi.org/10.3390/ijms21238991
Rashidieh B (2019) Deciphering the functional roles of centrosomal protein 55 (CEP55) in development and cancer: school of environment and science. Griffith Univ. https://doi.org/10.25904/1912/3496
Buzun K, Gornowicz A, Lesyk R, Bielawski K, Bielawska AJIJoMS (2021) Autophagy modulators in cancer therapy. Int J Mol Sci 22(11):5804. https://doi.org/10.3390/ijms22115804
Acknowledgements
The authors acknowledge BITS Pilani, Pilani campus, for providing them with infrastructural support. Heena Saini thanks CSIR for providing a student fellowship.
Funding
We thank DST-SERB Fast Track Project of RC (Sanction order No. SB/FT/LS-233/2012), DBT-Pilot Project of RC (BT/PR/8799/MED/30/1067/2013), ICMR of RC (2020-1404/ADHOC-BMS) and SERB of RC (EMR/2016/006788) for providing funding support for conducting the experiments.
Author information
Authors and Affiliations
Contributions
HS-I performed and analyzed the experiments, MC helped with wet-lab experiments and revision, HS-I, HS-II and SC performed the in silico and statistical analysis; SM helped in experimental analysis and manuscript correction; RC received funding for research; HS-I and RC designed the study plan and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIF 5891.1 kb)
Supplementary Fig. 1. A Immunoblot post siRNA mediated ablation of P53 in R273H-P53-H1299 cells (n = 3, p < 0.0001). ‘Cntrl’ stands for scrambled siRNA. GAPDH served as loading control. B Transcript levels of p53 (n = 3, p = 0.0361), upon over-expression of YAP (YAP-OE) in SW480 cells. ‘∗’ indicates a significant difference compared to empty vector (EV) transfected control. C Immunoblot representing level of P53, upon overexpression of YAP (YAP-OE) in GOF-R156P-P53 harboring HOS cells. Actin served as loading control. D Cell viability, as measured through MTT assay after exposure to pifithrin-alpha (PFT-α) [n = 3; p > 0.05 (24 hours) and p < 0.001 (48, 72 and 96 hours) at 10 µM dose. ‘∗’ indicates a significant difference compared to untreated cells (Cntrl); one way ANOVA (vs Time). E Graph showing relation between expression of autophagy associated genes and 5-year survival (%) of NSCLC patients. Gene expression data analyzed from GDC portal. F Difference in LC3B-II protein expression between P53-Null and R273H-P53 expressing H1299 cells (n = 3, p = 0.0012) and immunoblot of P53 protein in H1299 null and H1299-R273H cells. GAPDH served as loading control. In the figures, wherever applicable, bar graphs indicate mean ± standard error of mean (SEM). Unless otherwise mentioned, T-test was used for analysis of statistical significance. The statistically significant differences in comparison to control are represented with * and their corresponding p values are given below: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001
Supplementary material 2 (TIF 30756.5 kb)
Supplementary Fig. 2. A The fluorescence intensity profile across the white line as analyzed from microscopic image for both green and red channels is shown in the graph representing YAP and R273H-P53 co-localization. B Immuno-fluorescence image showing YAP (Red) and R273H-P53 (Green) localization 48 h after si-ATG-5 transfection or C Baf (10 nM) treatment in R273H-P53 harboring H1299 cells. D Immuno-fluorescence image showing YAP localization post CQ treatment (50 µM) for 48 h in P53-Null H1299 cells. Scale bar: 10 μm. Objective: Plan Apochromat 63x/1.40 oil M27. E Immuno-fluorescence image showing YAP and mutant-P53 localization post CQ treatment (50 µM) for 48 h in HOS cells. DAPI (Blue) was used to stain the nucleus. T-test was used for analysis of statistical significance. The statistically significant differences in comparison to control are represented with * and their corresponding p values are given below: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001
Supplementary material 3 (TIF 4964.3 kb)
Supplementary Fig. 3. The fluorescence intensity profile across the white line as analyzed from microscopic images for both green and red channels is shown in the graphs representing protein co-localization. A The fluorescence intensity profiles for YAP and LTR co-localization (one way ANOVA, vs CQ); B YAP and LAMP-2a co-localization; C R273H-P53 and LAMP-2a co-localization; D HSC-70 and YAP co-localization post CQ exposure (50 µM) for 48 hours. In the figures, ‘Cntrl’ stands for untreated cells. The statistically significant differences in comparison to control/CQ are represented with */# and their corresponding p values are given below: */#p ≤ 0.05, **/## p ≤ 0.01, ***/###p ≤ 0.001
Supplementary material 4 (TIF 24634.8 kb)
Supplementary Fig. 4. A SEM images showing comparative cytoplasmic changes in cells cultured in CQ-free media (withdrawal; W). B Immuno-fluorescence image of YAP and R273H-P53 post CQ withdrawal. Scale bar: 10 µm. Objective: Plan Apochromat 63x/1.40 oil M27. White arrows indicate accumulation and co-localization. ‘∗’ or ‘#’ indicates a significant difference compared to untreated or CQ treated cells respectively. C Transcript levels of YAP (n = 4, p = 0.0426) and CYR61 (n = 3, p = 0.0106) after CQ withdrawal. CQ treatment (50 µM) was for 48 hours; however, for withdrawal experiments cells were exposed to CQ for 24 hours followed by culture in CQ free media for 24 hours. ‘#’ indicates a significant difference compared to CQ treated cells. In the figures, unless otherwise mentioned, T-test was used for analysis of statistical significance. The statistically significant differences in comparison to control/CQ are represented with */# and their corresponding p values are given below: */#p ≤ 0.05, **/##p ≤ 0.01, ***/###p ≤ 0.001.
Supplementary material 5 (TIF 7573.4 kb)
Supplementary Fig. 5. A schematic diagram representing the overall effect of CQ exposure as observed in our study
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saini, H., Choudhary, M., Sharma, H. et al. Chloroquine induces transitory attenuation of proliferation of human lung cancer cells through regulation of mutant P53 and YAP. Mol Biol Rep 50, 1045–1058 (2023). https://doi.org/10.1007/s11033-022-08072-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08072-y