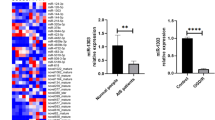
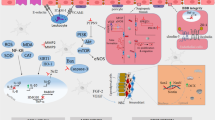
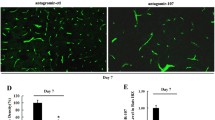
Abstract
Ischemic stroke accounts for about 71% of strokes worldwide. Due to limited recommended therapeutics for ischemic stroke, more attention is focused on angiogenesis in ischemic stroke. Not long after ischemic stroke, angiogenesis arises and is vital for the prognosis. Various pro-angiogenic, anti-angiogenic factors and their downstream pathways engage in angiogenesis regulation. CircRNAs are differentially expressed after ischemic stroke. Up to now, circRNAs have been found to exert many functions in regulating apoptosis, autophagy, proliferation, and differentiation of neurons and neural stem cells mainly as miRNAs sponges or proteins decoy. Thus, many circRNAs are considered promising biomarkers or therapeutic targets for ischemic stroke. Besides, circRNAs participate in the modulation of endothelial-mesenchymal transition and blood-brain barrier maintenance. Moreover, circRNAs play significant roles in endothelial dysfunction concerning inflammation responses, apoptosis, proliferation, and migration. They correlate with many angiogenesis-related signaling pathways and genes via the circRNA/miRNA/mRNA network. Novel insights into circRNAs significance in angiogenesis regulation in ischemic stroke could be provided for further researches on the clinical application of circRNAs in ischemic stroke.

Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- IS:
-
Ischemic stroke
- VECs:
-
Vascular endothelial cells
- MMPs:
-
Matrix metalloproteinases
- FGF:
-
Fibroblast growth factors
- TIMP:
-
Tissue inhibitor of matrix metalloproteinase
- GPCRs:
-
G protein-coupled receptors
- CSF:
-
Granulocyte colony-stimulating factor
- BBB:
-
Blood-brain barrier
- ICSs:
-
Intronic complementary sequences
- RBPs:
-
RNA binding proteins
- EcircRNAs:
-
Exonic circular RNAs
- CiRNAs:
-
Intronic circular RNAs
- EIciRNAs:
-
Exon–intron circular RNAs
- MBL/MBNL1:
-
Muscleblind
- SEP3:
-
SEPALLATA3
- SRSF1:
-
Serine/Arginine rich splicing factor 1
- m6A:
-
N6-Methyladenosine
- IRESs:
-
Internal ribosome entry sites
- MIRESs m6A:
-
Induced ribosome engagement sites
- BMECs:
-
Brain microvascular endothelial cells
- HBMECs:
-
Human brain microvascular endothelial cells
- OGD/R :
-
Oxygen-glucose deprivation/reperfusion
- HUVEC:
-
Human umbilical vein endothelial cell
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DE:
-
Differentially expressed
- DLK1:
-
Delta-like 1
- ATG5:
-
Autophagy-related 5
- TJPs:
-
Tight junction proteins
- EndoMT:
-
Endothelial-mesenchymal transition
- DNMT1:
-
DNA methyltransferase 1
- SOCS3:
-
Suppressor of cytokine signaling 3
- JNK:
-
c-Jun N terminal kinase;
- STAT3:
-
Signal transducer and activator of transcription 3
- NF-kβ:
-
Nuclear factor kappa-B
- HECTD1:
-
HECT domain E3 ubiquitin protein ligase 1
- TRAF3:
-
Tumor necrosis factor receptor-associated factor 3
- VEZF1:
-
Vascular endothelial zinc finger 1
- SOD2:
-
Superoxide dismutase 2
- NR4A1:
-
Nuclear receptor subfamily 4 group A member 1
- PTEN:
-
Phosphatase and tensin homolog deleted on chromosome ten
References
Katan M, Luft A (2018) Global Burden of Stroke. Semin Neurol 38:208–211. https://doi.org/10.1055/s-0038-1649503
Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A et al (2013) Council on E, Prevention, Council on Peripheral Vascular D, Council on Nutrition PA and An updated definition of stroke for the 21st century: a statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke 44:2064-89. https://doi.org/10.1161/STR.0b013e318296aeca
Meschia JF, Brott T (2018) Ischaemic stroke. Eur J Neurol 25:35–40. https://doi.org/10.1111/ene.13409
Smith M, Reddy U, Robba C, Sharma D, Citerio G (2019) Acute ischaemic stroke: challenges for the intensivist. Intensive Care Med 45:1177–1189. https://doi.org/10.1007/s00134-019-05705-y
Chavez LM, Huang SS, MacDonald I, Lin JG, Lee YC, Chen YH (2017) Mechanisms of Acupuncture Therapy in Ischemic Stroke Rehabilitation: A Literature Review of Basic Studies. Int J Mol Sci 18. https://doi.org/10.3390/ijms18112270
Herpich F, Rincon F (2020) Management of Acute Ischemic Stroke. Crit Care Med 48:1654–1663. https://doi.org/10.1097/CCM.0000000000004597
Powers WJ, Rabinstein AA, Ackerson T, Adeoye OM, Bambakidis NC, Becker K et al (2019) Guidelines for the Early Management of Patients With Acute Ischemic Stroke: 2019 Update to the 2018 Guidelines for the Early Management of Acute Ischemic Stroke: A Guideline for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke 50:e344–e418. https://doi.org/10.1161/STR.0000000000000211
Kir D, Schnettler E, Modi S, Ramakrishnan S (2018) Regulation of angiogenesis by microRNAs in cardiovascular diseases. Angiogenesis 21:699–710. https://doi.org/10.1007/s10456-018-9632-7
Hatakeyama M, Ninomiya I, Kanazawa M (2020) Angiogenesis and neuronal remodeling after ischemic stroke. Neural Regen Res 15:16–19. https://doi.org/10.4103/1673-5374.264442
Hayashi T, Noshita N, Sugawara T, Chan PH (2003) Temporal profile of angiogenesis and expression of related genes in the brain after ischemia. J Cereb Blood Flow Metab 23:166–180. https://doi.org/10.1097/01.WCB.0000041283.53351.CB
Krupinski J, Kaluza J, Kumar P, Kumar S, Wang JM (1994) Role of angiogenesis in patients with cerebral ischemic stroke. Stroke 25:1794–1798. https://doi.org/10.1161/01.str.25.9.1794
Szpak GM, Lechowicz W, Lewandowska E, Bertrand E, Wierzba-Bobrowicz T, Dymecki J (1999) Border zone neovascularization in cerebral ischemic infarct. Folia Neuropathol 37:264–268
Li G, Yu F, Lei T, Gao H, Li P, Sun Y et al (2016) Bone marrow mesenchymal stem cell therapy in ischemic stroke: mechanisms of action and treatment optimization strategies. Neural Regen Res 11:1015–1024. https://doi.org/10.4103/1673-5374.184506
Arenillas JF, Sobrino T, Castillo J, Davalos A (2007) The role of angiogenesis in damage and recovery from ischemic stroke. Curr Treat Options Cardiovasc Med 9:205–212. https://doi.org/10.1007/s11936-007-0014-5
Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W (2020) CircRNA: a rising star in gastric cancer. Cell Mol Life Sci 77:1661–1680. https://doi.org/10.1007/s00018-019-03345-5
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y (2016) Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 44:1370–1383. https://doi.org/10.1093/nar/gkv1367
Li X, Yang L, Chen LL (2018) The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 71:428–442. https://doi.org/10.1016/j.molcel.2018.06.034
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P et al (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–1030. https://doi.org/10.1016/0092-8674(93)90279-y
Viallard C, Larrivee B (2017) Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis 20:409–426. https://doi.org/10.1007/s10456-017-9562-9
Nowak-Sliwinska P, Alitalo K, Allen E, Anisimov A, Aplin AC, Auerbach R et al (2018) Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis 21:425–532. https://doi.org/10.1007/s10456-018-9613-x
Chandra A, Rick J, Yagnik G, Aghi MK (2020) Autophagy as a mechanism for anti-angiogenic therapy resistance. Semin Cancer Biol 66:75–88. https://doi.org/10.1016/j.semcancer.2019.08.031
Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR (2019) Vessel co-option in cancer. Nat Rev Clin Oncol 16:469–493. https://doi.org/10.1038/s41571-019-0181-9
Bikfalvi A (2017) History and conceptual developments in vascular biology and angiogenesis research: a personal view. Angiogenesis 20:463–478. https://doi.org/10.1007/s10456-017-9569-2
Zhang TR, Huang WQ (2020) Angiogenic circular RNAs: A new landscape in cardiovascular diseases. Microvasc Res 129:103983. https://doi.org/10.1016/j.mvr.2020.103983
Zhang W, Wu Y, Chen H, Yu D, Zhao J, Chen J (2021) Neuroprotective effects of SOX5 against ischemic stroke by regulating VEGF/PI3K/AKT pathway. Gene 767:145148. https://doi.org/10.1016/j.gene.2020.145148
Katoh M (2013) Therapeutics targeting angiogenesis: genetics and epigenetics, extracellular miRNAs and signaling networks (Review). Int J Mol Med 32:763–767. https://doi.org/10.3892/ijmm.2013.1444
Akwii RG, Sajib MS, Zahra FT, Mikelis CM (2019) Role of Angiopoietin-2 in Vascular Physiology and Pathophysiology. Cells 8. https://doi.org/10.3390/cells8050471
Quintero-Fabian S, Arreola R, Becerril-Villanueva E, Torres-Romero JC, Arana-Argaez V, Lara-Riegos J et al (2019) Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front Oncol 9:1370. https://doi.org/10.3389/fonc.2019.01370
Katoh M (2016) Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol Sci 37:1081–1096. https://doi.org/10.1016/j.tips.2016.10.003
Jiang S, Fu R, Shi J, Wu H, Mai J, Hua X et al (2021) CircRNA-Mediated Regulation of Angiogenesis: A New Chapter in Cancer Biology. Front Oncol 11:553706. https://doi.org/10.3389/fonc.2021.553706
Guo L, Xu B, Zhou D, Chang G, Fu Y, Liu L, Luo Y (2019) Biophysical and biological characterization of PEGylated recombinant human endostatin. Clin Exp Pharmacol Physiol 46:920–927. https://doi.org/10.1111/1440-1681.13134
Song Q, Ji Q, Li Q (2018) The role and mechanism of betaarrestins in cancer invasion and metastasis (Review). Int J Mol Med 41:631–639. https://doi.org/10.3892/ijmm.2017.3288
Shirzad R, Shahrabi S, Ahmadzadeh A, Kampen KR, Shahjahani M, Saki N (2016) Signaling and molecular basis of bone marrow niche angiogenesis in leukemia. Clin Transl Oncol 18:957–971. https://doi.org/10.1007/s12094-015-1477-6
Venugopal S, Kao C, Chandna R, Sulochana KN, Subramanian V, Chen M et al (2018) Angio-3, a 10-residue peptide derived from human plasminogen kringle 3, suppresses tumor growth in mice via impeding both angiogenesis and vascular permeability. Angiogenesis 21:653–665. https://doi.org/10.1007/s10456-018-9616-7
Liu J, Zhou X, Li Q, Zhou SM, Hu B, Hu GW et al (2017) Role of Phosphorylated HDAC4 in Stroke-Induced Angiogenesis. Biomed Res Int 2017:2957538. https://doi.org/10.1155/2017/2957538
Gandin C, Widmann C, Lazdunski M, Heurteaux C (2016) MLC901 Favors Angiogenesis and Associated Recovery after Ischemic Stroke in Mice. Cerebrovasc Dis 42:139–154. https://doi.org/10.1159/000444810
Chen J, Zhang X, Liu X, Zhang C, Shang W, Xue J et al (2019) Ginsenoside Rg1 promotes cerebral angiogenesis via the PI3K/Akt/mTOR signaling pathway in ischemic mice. Eur J Pharmacol 856:172418. https://doi.org/10.1016/j.ejphar.2019.172418
Lee ST, Chu K, Jung KH, Ko SY, Kim EH, Sinn DI et al (2005) Granulocyte colony-stimulating factor enhances angiogenesis after focal cerebral ischemia. Brain Res 1058:120–128. https://doi.org/10.1016/j.brainres.2005.07.076
Zan L, Wu H, Jiang J, Zhao S, Song Y, Teng G et al (2011) Temporal profile of Src, SSeCKS, and angiogenic factors after focal cerebral ischemia: correlations with angiogenesis and cerebral edema. Neurochem Int 58:872–879. https://doi.org/10.1016/j.neuint.2011.02.014
Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH et al (2003) SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med 9:900–906. https://doi.org/10.1038/nm889
Lu S, Yang X, Wang C, Chen S, Lu S, Yan W et al (2019) Current status and potential role of circular RNAs in neurological disorders. J Neurochem 150:237–248. https://doi.org/10.1111/jnc.14724
Yang J, Chen M, Cao RY, Li Q, Zhu F (2018) The Role of Circular RNAs in Cerebral Ischemic Diseases: Ischemic Stroke and Cerebral Ischemia/Reperfusion Injury. Adv Exp Med Biol 1087:309–325. https://doi.org/10.1007/978-981-13-1426-1_25
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR et al (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177. https://doi.org/10.1016/j.celrep.2014.12.019
Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29:2168–2182. https://doi.org/10.1101/gad.270421.115
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Zaphiropoulos PG (1996) Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 93:6536–6541. https://doi.org/10.1073/pnas.93.13.6536
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. https://doi.org/10.1016/j.molcel.2013.08.017
Ebbesen KK, Hansen TB, Kjems J (2017) Insights into circular RNA biology. RNA Biol 14:1035–1045. https://doi.org/10.1080/15476286.2016.1271524
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J et al (2017) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 18:984–992. https://doi.org/10.1093/bib/bbw081
Hernandez-Romero IA, Guerra-Calderas L, Salgado-Albarran M, Maldonado-Huerta T, Soto-Reyes E (2019) The Regulatory Roles of Non-coding RNAs in Angiogenesis and Neovascularization From an Epigenetic Perspective. Front Oncol 9:1091. https://doi.org/10.3389/fonc.2019.01091
Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL, Guo SS (2020) CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep 10:9024. https://doi.org/10.1038/s41598-020-65920-2
Zhang N, Gao Y, Yu S, Sun X, Shen K (2020) Berberine attenuates Abeta42-induced neuronal damage through regulating circHDAC9/miR-142-5p axis in human neuronal cells. Life Sci 252:117637. https://doi.org/10.1016/j.lfs.2020.117637
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691. https://doi.org/10.1038/s41576-019-0158-7
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. https://doi.org/10.1038/nsmb.2959
Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G et al (2017) A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 3:17053. https://doi.org/10.1038/nplants.2017.53
Barbagallo D, Caponnetto A, Barbagallo C, Battaglia R, Mirabella F, Brex D et al (2021) The GAUGAA Motif Is Responsible for the Binding between circSMARCA5 and SRSF1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Int J Mol Sci 22. https://doi.org/10.3390/ijms22041678
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14:361–369. https://doi.org/10.1080/15476286.2017.1279788
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W et al (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429. https://doi.org/10.1038/ncomms12429
Li B, Zhu L, Lu C, Wang C, Wang H, Jin H et al (2021) circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun 12:295. https://doi.org/10.1038/s41467-020-20527-z
Huang S, Li X, Zheng H, Si X, Li B, Wei G et al (2019) Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 139:2857–2876. https://doi.org/10.1161/CIRCULATIONAHA.118.038361
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z et al (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412. https://doi.org/10.1093/eurheartj/ehw001
Prats AC, David F, Diallo LH, Roussel E, Tatin F, Garmy-Susini B, Lacazette E (2020) Circular RNA, the Key for Translation. Int J Mol Sci 21. https://doi.org/10.3390/ijms21228591
Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC et al (2019) How are circRNAs translated by non-canonical initiation mechanisms? Biochimie 164:45–52. https://doi.org/10.1016/j.biochi.2019.06.015
Yang Y, Wang Z (2019) IRES-mediated cap-independent translation, a path leading to hidden proteome. J Mol Cell Biol 11:911–919. https://doi.org/10.1093/jmcb/mjz091
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O et al (2017) Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 66:22–37e9. https://doi.org/10.1016/j.molcel.2017.02.017
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L et al (2017) Translation of CircRNAs. Mol Cell 66:9–21e7. https://doi.org/10.1016/j.molcel.2017.02.021
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F et al (2018) Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 110. https://doi.org/10.1093/jnci/djx166
Begum S, Yiu A, Stebbing J, Castellano L (2018) Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene 37:4055–4057. https://doi.org/10.1038/s41388-018-0230-3
Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST et al (2019) Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 20:84. https://doi.org/10.1186/s13059-019-1685-4
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P et al (2018) A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9:4475. https://doi.org/10.1038/s41467-018-06862-2
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H et al (2019) A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer 18:131. https://doi.org/10.1186/s12943-019-1056-5
Fazi F, Fatica A (2019) Interplay Between N (6)-Methyladenosine (m(6)A) and Non-coding RNAs in Cell Development and Cancer. Front Cell Dev Biol 7:116. https://doi.org/10.3389/fcell.2019.00116
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y et al (2017) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27:626–641. https://doi.org/10.1038/cr.2017.31
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z (2020) The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer 19:105. https://doi.org/10.1186/s12943-020-01224-3
Cheetham SW, Faulkner GJ, Dinger ME (2020) Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat Rev Genet 21:191–201. https://doi.org/10.1038/s41576-019-0196-1
Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X et al (2012) Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149:1622–1634. https://doi.org/10.1016/j.cell.2012.04.041
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L (2016) CircRNA-derived pseudogenes. Cell Res 26:747–750. https://doi.org/10.1038/cr.2016.42
Chen L, Luo W, Zhang W, Chu H, Wang J, Dai X et al (2020) circDLPAG4/HECTD1 mediates ischaemia/reperfusion injury in endothelial cells via ER stress. RNA Biol 17:240–253. https://doi.org/10.1080/15476286.2019.1676114
Gan L, Liao S, Xing Y, Deng S (2020) The Regulatory Functions of lncRNAs on Angiogenesis Following Ischemic Stroke. Front Mol Neurosci 13:613976. https://doi.org/10.3389/fnmol.2020.613976
Wen Y, Chun Y, Lian ZQ, Yong ZW, Lan YM, Huan L et al (2021) circRNA–0006896–miR1264–DNMT1 axis plays an important role in carotid plaque destabilization by regulating the behavior of endothelial cells in atherosclerosis. Molecular medicine reports 23. https://doi.org/10.3892/mmr.2021.11950
Jiang S, Zhao G, Lu J, Jiang M, Wu Z, Huang Y et al (2020) Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol Res 53:27. https://doi.org/10.1186/s40659-020-00295-2
Li J, Wang J, Wang Z (2021) Circ_0006768 upregulation attenuates oxygen-glucose deprivation/reoxygenation-induced human brain microvascular endothelial cell injuries by upregulating VEZF1 via miR-222-3p inhibition. Metab Brain Dis 36:2521–2534. https://doi.org/10.1007/s11011-021-00775-8
Sun X, Dai M, Liu X, Wang H, Wang C, Fan X, Fang W (2022) Hsa_circ_0090002 regulates miR-186-5p/HECTD1 axis to mediate brain microvascular endothelial cell dysfunction. Brain Res Bull 178:97–107. https://doi.org/10.1016/j.brainresbull.2021.11.007
Wang X, Liu L, Zhang L, Guo J, Yu L, Li T (2022) Circ_0057583 facilitates brain microvascular endothelial cell injury through modulating miR-204-5p/NR4A1 axis. Metab Brain Dis 37:501–511. https://doi.org/10.1007/s11011-021-00866-6
Yang X, Li X, Zhong C, Peng J, Pang J, Peng T et al (2021) Circular RNA circPHKA2 Relieves OGD-Induced Human Brain Microvascular Endothelial Cell Injuries through Competitively Binding miR-574-5p to Modulate SOD2. Oxid Med Cell Longev 2021:3823122. https://doi.org/10.1155/2021/3823122
Xu X, Wu Z, Qiu H, Wu J (2021) Circular RNA circPHC3 Promotes Cell Death and Apoptosis in Human BMECs After Oxygen Glucose Deprivation via miR-455-5p/TRAF3 Axis in vitro. Neuropsychiatr Dis Treat 17:147–156. https://doi.org/10.2147/NDT.S288669
Bai X, Liu X, Wu H, Feng J, Chen H, Zhou D (2022) CircFUNDC1 knockdown alleviates oxygen-glucose deprivation-induced human brain microvascular endothelial cell injuries by inhibiting PTEN via miR-375. Neurosci Lett 770:136381. https://doi.org/10.1016/j.neulet.2021.136381
Li S, Chen L, Xu C, Qu X, Qin Z, Gao J et al (2020) Expression profile and bioinformatics analysis of circular RNAs in acute ischemic stroke in a South Chinese Han population. Sci Rep 10:10138. https://doi.org/10.1038/s41598-020-66990-y
Li F, Li C, Li X, Li Y, Zhong Y, Ling L (2020) Altered circular RNA expression profiles in the non-ischemic thalamus in focal cortical infarction mice. Aging 12:13206–13219. https://doi.org/10.18632/aging.103424
Jia L, Zhou X, Huang X, Xu X, Jia Y, Wu Y et al (2018) Maternal and umbilical cord serum-derived exosomes enhance endothelial cell proliferation and migration. FASEB J 32:4534–4543. https://doi.org/10.1096/fj.201701337RR
Li X, Zhang Y, Wang Y, Zhao D, Sun C, Zhou S et al (2020) Exosomes Derived from CXCR4-Overexpressing BMSC Promoted Activation of Microvascular Endothelial Cells in Cerebral Ischemia/Reperfusion Injury. Neural Plast 2020:8814239. https://doi.org/10.1155/2020/8814239
Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, Barres BA (2009) Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci U S A 106:641–646. https://doi.org/10.1073/pnas.0805165106
Yan G, Zhao H, Hong X (2020) LncRNA MACC1-AS1 attenuates microvascular endothelial cell injury and promotes angiogenesis under hypoxic conditions via modulating miR-6867-5p/TWIST1 in human brain microvascular endothelial cells. Ann Transl Med 8:876. https://doi.org/10.21037/atm-20-4915
Liu W, Jia C, Luo L, Wang HL, Min XL, Xu JH et al (2019) Novel circular RNAs expressed in brain microvascular endothelial cells after oxygen-glucose deprivation/recovery. Neural Regen Res 14:2104–2111. https://doi.org/10.4103/1673-5374.262589
Wu L, Xu H, Zhang W, Chen Z, Li W, Ke W (2020) Circular RNA circCCDC9 alleviates ischaemic stroke ischaemia/reperfusion injury via the Notch pathway. J Cell Mol Med 24:14152–14159. https://doi.org/10.1111/jcmm.16025
Park S, Sorenson CM, Sheibani N (2015) PECAM-1 isoforms, eNOS and endoglin axis in regulation of angiogenesis. Clin Sci (Lond) 129:217–234. https://doi.org/10.1042/CS20140714
Huang CC, Kuo HM, Wu PC, Cheng SH, Chang TT, Chang YC et al (2018) Soluble delta-like 1 homolog (DLK1) stimulates angiogenesis through Notch1/Akt/eNOS signaling in endothelial cells. Angiogenesis 21:299–312. https://doi.org/10.1007/s10456-018-9596-7
Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu G, Yao H (2018) Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy 14:404–418. https://doi.org/10.1080/15548627.2017.1414755
Sprott D, Poitz DM, Korovina I, Ziogas A, Phieler J, Chatzigeorgiou A et al (2019) Endothelial-Specific Deficiency of ATG5 (Autophagy Protein 5) Attenuates Ischemia-Related Angiogenesis. Arterioscler Thromb Vasc Biol 39:1137–1148. https://doi.org/10.1161/ATVBAHA.119.309973
Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R et al (2018) Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J Neurosci 38:32–50. https://doi.org/10.1523/JNEUROSCI.1348-17.2017
Funding
This This work was funded by the Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital, Grant No. 2020LNJJ16), the Provincial Key Plan for Research and Development of Hunan (Grant No. 2020SK2067; No. 2020SK2069), the Natural Science Foundation of Hunan Province (Grant No. 2021JJ31109; No. 2020JJ4875), and the Fundamental Research Funds for the Central Universities of Central South University [Grant No. 2021zzts1029; No. 2020zzts269].
Author information
Authors and Affiliations
Contributions
The authors declare that all data were generated in-house and that no paper mill was used. Jian Xia contributes to conception. Liuyang Cheng made contributions in retrieving literature, collecting information, and writing the manuscript. Zeyu Liu and Jian Xia participate in revising it and give critical intellectual suggestions.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, L., Liu, Z. & Xia, J. New insights into circRNA and its mechanisms in angiogenesis regulation in ischemic stroke: a biomarker and therapeutic target. Mol Biol Rep 50, 829–840 (2023). https://doi.org/10.1007/s11033-022-07949-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07949-2