Abstract
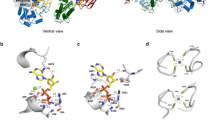
In the present investigation, we report cloning, expression, purification and characterization of a novel Bleomycin Resistance Dioxygenase (BRPD). His-tagged fusion protein was purified to homogeneity using Ni-NTA affinity chromatography, yielding 1.2 mg of BRPD with specific activity of 6.25 U mg−1 from 600 ml of E. coli culture. Purified enzyme was a dimer with molecular weight ~ 26 kDa in SDS-PAGE and ~ 73 kDa in native PAGE analysis. The protein catalyzed breakdown of hydrocarbon substrates, including catechol and hydroquinone, in the presence of metal ions, as characterized via spectrophotometric analysis of the enzymatic reactions. Bleomycin binding was proven using the EMSA gel retardation assay, and the putative bleomycin binding site was further determined by in silico analysis. Molecular dynamic simulations revealed that BRPD attains octahedral configuration in the presence of Fe2+ ion, forming six co-ordinate complexes to degrade hydroquinone-like molecules. In contrary, in the presence of Zn2+ ion BRPD adopts tetrahedral configuration, which enables degradation of catechol-like molecules.






Similar content being viewed by others
References
Zhang T, Wooster MJ, Green DC, Main B (2015) New field-based agricultural biomass burning trace gas, PM 2.5, and black carbon emission ratios and factors measured in situ at crop residue fires in Eastern China. Atmos Environ 121:22–34
Fu B, Xu T, Cui Z, Ng HL, Wang K, Li J, Li QX (2018) Mutation of phenylalanine-223 to leucine enhances transformation of benzo[a]pyrene by ring-hydroxylating dioxygenase of Sphingobium sp. FB3 by increasing accessibility of the catalytic site. J Agric Food Chem 66(5):1206–1213
Santos DF, Istvan P, Noronha EF, Quirino BF, Krüger RH (2015) New dioxygenase from metagenomic library from Brazilian soil: insights into antibiotic resistance and bioremediation. Biotechnol Lett 37(9):1809–1817
Arora PK, Kumar M, Chauhan A, Raghava GP, Jain RK (2009) OxDBase: a database of oxygenases involved in biodegradation. BMC Res Notes 2:67
Cole JR, Konstantinidis K, Farris RJ, Tiedje JM (2010) Microbial diversity and phylogeny: extending from rRNAs to genomes. In: Liu W-T, Jansson JK (eds) Environmental molecular microbiology. Caister Academic Press, Norfolk, pp 1–19
Spain JC, Gibson DT (1991) Pathway for biodegradation of para-nitrophenol in a Moraxella sp. Appl Environ Microbiol 57(3):812–819
Zhang S, Sun W, Xu L, Zheng X, Chu X, Tian J, Wu N, Fan Y (2012) Identification of the para-nitrophenol catabolic pathway, and characterization of three enzymes involved in the hydroquinone pathway, in Pseudomonas sp. 1-7. BMC Microbiol 12:27
Miyauchi K, Adachi Y, Nagata Y, Takagi M (1999) Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of gamma-hexachlorocyclohexane in Sphingomonas paucimobilis. J Bacteriol 181(21):6712–6719
Moonen MJH, Kamerbeek NM, Westphal AH, Boeren SA, Janssen DB, Fraaije MW, van Berkel WJH (2008) Elucidation of the 4 hydroxyacetophenone catabolic pathway in Pseudomonas fluorescens ACB. J Bacteriol 190(15):5190–5198
Takenaka S, Okugawa S, Kadowaki M, Murakami S, Aoki K (2003) The metabolic pathway of 4-aminophenol in Burkholderia sp strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl Environ Microbiol 69(9):5410–5413
Kolvenbach BA, Lenz M, Benndorf D, Rapp E, Fousek J, Vlcek C, Schäffer A, Gabriel FL, Kohler HP, Corvini PF (2011) Purification and characterization of hydroquinone dioxygenase from Sphingomonas sp. strain TTNP3. AMB Express 1(1):8
Eltis LD, Bolin JT (1996) Evolutionary relationships among extradiol dioxygenases. J Bacteriol 178(20):5930–5937
Hayes RP, Green AR, Nissen MS, Lewis KM, Xun L, Kang C (2013) Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme. Mol Microbiol 88(3):523–536
He P, Moran G (2011) Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J Inorg Biochem 105(10):1259–1272
Machonkin TE, Holland PL, Smith KN, Liberman JS, Dinescu A, Cundari TR, Rocks SS (2010) Determination of the active site of Sphingobium chlorophenolicum 2,6-dichlorohydroquinone dioxygenase (PcpA). J Biol Inorg Chem 15(3):291–300
McCarthy AA, Baker HM, Shewry SC, Patchett ML, Baker EN (2001) Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure 9(7):637–646
Dortet L, Girlich D, Virlouvet AL, Poirel L, Nordmann P, Iorga BI, Naas T (2017) Characterization of BRPMBL, the bleomycin resistance protein associated with the carbapenemase NDM. Antimicrob Agents Chemother 61(3):2413–2416
Zhang JJ, Liu H, Xiao Y, Zhang XE, Zhou NY (2009) Identification and characterization of catabolic para-Nitrophenol 4-Monooxygenase and para- benzoquinone reductase from Pseudomonas sp. strain WBC-3. J Bacteriol 191(8):2703–2710
Shen W, Liu W, Zhang J, Tao J, Deng H, Cao H, Cui Z (2010) Cloning and characterization of a gene cluster involved in the catabolism of p-nitrophenol from Pseudomonas putida DLL-E4. Bioresour Technol 101(19):7516–7522
Min J, Zhang JJ, Zhou NY (2014) The gene cluster for para-Nitrophenol catabolism is responsible for 2-Chloro-4-Nitrophenol degradation in Burkholderia sp. strain SJ98. Appl Environ Microbiol 80(19):6212–6222
Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65(1):7–17
Armstrong RN (2000) Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39(45):13625–13632
Yin Y, Zhou NY (2010) Characterization of MnpC, a hydroquinone dioxygenase likely involved in the meta-nitrophenol degradation by Cupriavidus necator JMP134. Curr Microbiol 61(5):471–476
Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinform 86:2.9.1–2.9.37
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1):7–8
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44(W1):W344–W350
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38(Web Server issue):W529–W533
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11(8):3696–3713
Jorgensen WL, Chandrasekhar J, Madura JD (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Comput Phys 23(3):327
Kumar R, Shaikh N, Sharma S, Garg P (2015) A comparative study of integrase binding domain of homologous LEDGF/p75 and HRP2 protein: from sequence analysis to structural characterization. Mol Simul 41(8):683–690
Kumar R, Garg P (2013) Molecular modeling and active site binding mode characterization of aspartate β-semialdehyde dehydrogenase family. Mol Inform 32(4):377–383
Tropel D, Meyer C, Armengaud J, Jouanneau Y (2002) Ferredoxin-mediated reactivation of the chlorocatechol 2,3-dioxygenase from Pseudomonas putida GJ31. Arch Microbiol 177(4):345–351
Mancini S, Abicht HK, Gonskikh Y, Solioz M (2015) A copper-induced quinone degradation pathway provides protection against combined copper/quinone stress in Lactococcus lactis IL1403. Mol Microbiol 95(4):645–659
Cerdan P, Wasserfallen A, Rekik M, Timmis KN, Harayama S (1994) Substrates pecificity of catechol 2,3-dioxygenase encoded by tol plasmid pWWO of pseudomonas putida and its relationship to cell-growth. J Bacteriol 176(19):6074–6081
Bartels I, Knackmuss HJ, Reineke W (1984) Suicide inactivation of catechol 2,3- dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol 47(3):500–505
Ono K, Nozaki M, Hayaishi O (1970) Purification and some properties of protocatechuate 4,5-dioxygenase. Biochim et Biophys Acta 220(2):224–238
Guan Y, Zhu Q, Huang D, Zhao S, Jan Lo L, Peng J (2015) An equation to estimate the difference between theoretically predicted and SDS PAGE-displayed molecular weights for an acidic peptide. Sci Rep 5(1):13370
Bergdoll M, Eltis LD, Cameron AD, Dumas P, Bolin JT (1998) All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci 7(8):1661–1670
Jafari S, Kazemi N, Ryde U, Irani M (2018) Higher flexibility of Glu-172 explains the unusual stereospecificity of glyoxalase I. Inorg Chem 57(9):4944–4958
Cameron AD, Olin B, Ridderstrom M, Mannervik B, Jones TA (1997) Crystal structure of human glyoxalase I -evidence for gene duplication and 3D domain swapping. EMBO J 16(12):3386–3395
Fetzner S (2012) Ring-cleaving dioxygenases with a cupin fold. Appl Environ Microbiol 78(8):2505–2514
Schlunegger MP, Bennett MJ, Eisenberg D (1997) Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv Protein Chem 50:61–122
Saint-Jean AP, Phillips KR, Creighton DJ, Stone MJ (1998) Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: evidence for 3D domain swapping. Biochemistry 37(29):10345–10353
Funding
This research Grant was funded by the Science and Engineering Research Board (SERB) New Delhi, project file number: SB/YS/LS-63/2013 under fast track scheme for the young scientists. Dr PKS would like to thank SERB for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2019_5159_MOESM2_ESM.jpg
Supplementary material 2 (JPEG 160 kb) Supplementary Fig. S2 Multiple sequence alignment of BRPD and its homologues. The secondary structures of BRPD (QED41507) and two other putative dioxygenases (PDB 3OAJ and 1ZSW) were compared. α helices and β sheets were indicated by red and yellow highlighting respectively. Binding pocket and catalytic residues were indicated in bold. The Fe-co-ordinating residues in the BRPD were indicated with an asterisk
Rights and permissions
About this article
Cite this article
Sharma, V., Kumar, R., Sharma, V.K. et al. Expression, purification, characterization and in silico analysis of newly isolated hydrocarbon degrading bleomycin resistance dioxygenase. Mol Biol Rep 47, 533–544 (2020). https://doi.org/10.1007/s11033-019-05159-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05159-x