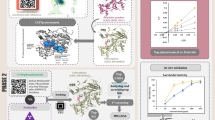
Abstract
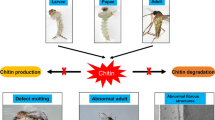
Throughout history, vector-borne diseases have consistently posed significant challenges to human health. Among the strategies for vector control, chemical insecticides have seen widespread use since their inception. Nevertheless, their effectiveness is continually undermined by the steady growth of insecticide resistance within these vector populations. As such, the demand for more robust, efficient, and cost-effective natural insecticides has become increasingly pressing. One promising avenue of research focuses on chitin, a crucial structural component of mosquitoes' exoskeletons and other insects. Chitin not only provides protection and rigidity but also lends flexibility to the insect body. It undergoes substantial transformations during insect molting, a process known as ecdysis. Crucially, the production of chitin is facilitated by an enzyme known as chitin synthase, making it an attractive target for potential novel insecticides. Our recent study delved into the impacts of curcumin, a natural derivative of turmeric, on chitin synthesis and larval development in Aedes aegypti, a mosquito species known to transmit dengue and yellow fever. Our findings demonstrate that even sub-lethal amounts of curcumin can significantly reduce overall chitin content and disrupt the cuticle development in the 4th instar larvae of Aedes aegypti. Further to this, we utilized computational analyses to investigate how curcumin interacts with chitin synthase. Techniques such as molecular docking, pharmacophore feature mapping, and molecular dynamics (MD) simulations helped to illustrate that curcumin binds to the same site as polyoxin D, a recognized inhibitor of chitin synthase. These findings point to curcumin's potential as a natural, bioactive larvicide that targets chitin synthase in mosquitoes and potentially other insects.
Graphical abstract













Similar content being viewed by others
Data availability
All the relevant data are contained within the manuscript. Additional raw data will be available upon request.
References
WHO (2020) Global Vector Control Response
Roth GA, Abate D, Abate KH et al (2018) Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1736–1788. https://doi.org/10.1016/S0140-6736(18)32203-7
Niang EHA, Bassene H, Fenollar F, Mediannikov O (2018) Biological control of mosquito-borne diseases: the potential of wolbachia-based interventions in an IVM framework. J Trop Med 14:8
Rocklöv J (2020) Dubrow R (2020) Climate change: an enduring challenge for vector-borne disease prevention and control. Nat Immunol 215(21):479–483. https://doi.org/10.1038/s41590-020-0648-y
Wilson AL, Courtenay O, Kelly-Hope LA et al (2020) The importance of vector control for the control and elimination of vector-borne diseases. PLoS Negl Trop Dis 14(1):e0007831
Khadka S, Proshad R, Thapa A et al (2020) (2020) Wolbachia : a possible weapon for controlling dengue in Nepal. Trop Med Heal 481(48):1–6. https://doi.org/10.1186/S41182-020-00237-4
van den Berg H, Velayudhan R, Yadav RS (2021) Management of insecticides for use in disease vector control: Lessons from six countries in Asia and the Middle East. PLoS Negl Trop Dis 15:e0009358. https://doi.org/10.1371/JOURNAL.PNTD.0009358
Correy GJ, Zaidman D, Harmelin A et al (2019) Overcoming insecticide resistance through computational inhibitor design. Proc Natl Acad Sci USA 116:21012–21021. https://doi.org/10.1073/pnas.1909130116
Rasoolizadeh A, Munger A, Goulet MC et al (2016) Functional proteomics-aided selection of protease inhibitors for herbivore insect control. Sci Rep. https://doi.org/10.1038/srep38827
Rao P, Goswami D, Rawal RM (2021) Revealing the molecular interplay of curcumin as Culex pipiens Acetylcholine esterase 1 (AChE1) inhibitor. Sci Rep 11:1–18. https://doi.org/10.1038/s41598-021-96963-8
Rao P, Goswami D, Rawal IDRM (2022) Extending the lore of curcumin as dipteran Butyrylcholine esterase (BChE) inhibitor: a holistic molecular interplay assessment. PLoS ONE 17:e0269036. https://doi.org/10.1371/JOURNAL.PONE.0269036
Rao P, Goswami D, Rawal RM (2022) Molecular insights on ar-turmerone as a structural, functional and pharmacophoric analogue of synthetic mosquito repellent DEET by comprehensive computational assessment. Sci Rep 121(12):1–17. https://doi.org/10.1038/s41598-022-19901-2
Muthukrishnan S, Merzendorfer H, Arakane Y, Kramer KJ (2012) Chitin metabolism in insects. Academic Press, Cambridge
Liu X, Cooper AMW, Yu Z et al (2019) Progress and prospects of arthropod chitin pathways and structures as targets for pest management. Pestic Biochem Physiol 161:33–46. https://doi.org/10.1016/J.PESTBP.2019.08.002
Merzendorfer H, Zimoch L (2003) Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206:4393–4412. https://doi.org/10.1242/JEB.00709
Zhu KY, Merzendorfer H, Zhang W et al (2016) Biosynthesis, turnover, and functions of chitin in insects. Annu Rev Entomol 61:177–196. https://doi.org/10.1146/annurev-ento-010715-023933
de Souza LM, Venturini FP, Inada NM et al (2020) Curcumin in formulations against Aedes aegypti: Mode of action, photolarvicidal and ovicidal activity. Photodiagnosis Photodyn Ther 31:101840. https://doi.org/10.1016/j.pdpdt.2020.101840
Matiadis D, Liggri PGV, Kritsi E et al (2021) Curcumin derivatives as potential mosquito larvicidal agents against two mosquito vectors, culex pipiens and aedes albopictus. Int J Mol Sci. https://doi.org/10.3390/ijms22168915
Jumper J, Evans R, Pritzel A et al (2021) (2021) Highly accurate protein structure prediction with AlphaFold. Nat 5967873(596):583–589. https://doi.org/10.1038/s41586-021-03819-2
Varadi M, Anyango S, Deshpande M et al (2022) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. https://doi.org/10.1093/NAR/GKAB1061
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28:235–242
Ren Z, Chhetri A, Guan Z et al (2022) Structural basis for inhibition and regulation of a chitin synthase from Candida albicans. Nat Struct Mol Biol 29:653–664. https://doi.org/10.1038/s41594-022-00791-x
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. https://doi.org/10.1093/bioinformatics/btq662
Chen VB, Arendall WB, Headd JJ et al (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D 66:12–21. https://doi.org/10.1107/S0907444909042073
Studer G, Rempfer C, Waterhouse AM et al (2020) QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 36:1765–1771. https://doi.org/10.1093/bioinformatics/btz828
Studer G, Biasini M, Schwede T (2014) Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane). Bioinformatics. https://doi.org/10.1093/bioinformatics/btu457
Schrödinger (2021) Schrödinger release 2021–2023: maestro. Schrödinger, LLC, New York
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
Shivakumar D, Williams J, Wu Y et al (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. ACS Publ 6:1509–1519. https://doi.org/10.1021/ct900587b
Kim S, Thiessen PA, Bolton EE et al (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. https://doi.org/10.1093/nar/gkv951
Greenwood JR, Calkins D, Sullivan AP, Shelley JC (2010) Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des 24:591–604
Shelley JC, Cholleti A, Frye LL et al (2007) Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21:681–691. https://doi.org/10.1007/s10822-007-9133-z
Friesner RA, Murphy RB, Repasky MP et al (2006) Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Kollman PA, Massova I, Reyes C et al (2000) Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. https://doi.org/10.1021/ar000033j
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM- PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des 18:113–135
Wang J, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided-Drug Des 2:287–306. https://doi.org/10.2174/157340906778226454
Wang W, Donini O, Reyes CM, Kollman PA (2001) Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu Rev Biophys Biomol Struct 30:211–243
Merzendorfer H (2011) The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur J Cell Biol 90:759–769. https://doi.org/10.1016/J.EJCB.2011.04.014
Muthukrishnan S, Merzendorfer H, Arakane Y, Yang Q (2016) Chitin metabolic pathways in insects and their regulation. Extracell Compos Matrices Arthropods. https://doi.org/10.1007/978-3-319-40740-1_2/COVER
Liu X, Cooper AMW, Zhang J, Zhu KY (2019) Biosynthesis, modifications and degradation of chitin in the formation and turnover of peritrophic matrix in insects. J Insect Physiol 114:109–115. https://doi.org/10.1016/J.JINSPHYS.2019.03.006
Wang Y, Gao L, Moussian B (2020) Drosophila, chitin and insect pest management. Curr Pharm Des 26:3546–3553. https://doi.org/10.2174/1381612826666200721002354
do Nascimento ARB, Pavinato VAC, Rodrigues JG, et al (2022) There is more than chitin synthase in insect resistance to benzoylureas: molecular markers associated with teflubenzuron resistance in Spodoptera frugiperda. J Pest Sci 2004(95):129–144. https://doi.org/10.1007/s10340-021-01373-4
Mohapatra R, Ranjit MR, Dash AP (1996) The effect of chitin synthesis inhibitors on the development of Brugia malayi in Aedes aegypti. J Helminthol 70:269–270. https://doi.org/10.1017/S0022149X00015522
Vasuki V (1992) Adult longevity of certain mosquito species after larval and pupal exposure to sublethal concentration of an insect growth regulator, hexaflumuron. Southeast Asian J Trop Med Public Health 23:121–124
Cooper CDO, Marsden BD (2017) N- and C-terminal truncations to enhance protein solubility and crystallization: predicting protein domain boundaries with bioinformatics tools. Methods Mol Biol 1586:11–31. https://doi.org/10.1007/978-1-4939-6887-9_2
van Eck WH (1979) Mode of action of two benzoylphenyl ureas as inhibitors of chitin synthesis in insects. Insect Biochem 9:295–300. https://doi.org/10.1016/0020-1790(79)90009-X
Ruiz-Herrera J, San-Blas G (2003) Chitin synthesis as a target for antifungal drugs. Curr Drug Targets Infect Disord 3:77–91. https://doi.org/10.2174/1568005033342064
Bowers B, Levin G, Cabib E (1974) Effect of Polyoxin D on Chitin synthesis and septum formation in saccharomyces cerevisiae. J Bacteriol 119:564–575. https://doi.org/10.1128/JB.119.2.564-575.1974
Funayama S, Isono K (2014) Biosynthesis of the polyoxins, nucleoside peptide antibiotics: formation of 5-carboxyuracil nucleosides by Streptomyces cacaoi. Agric Biol Chem 40:1039–1044. https://doi.org/10.1080/00021369197610862152
Dernoeden P (2012) Polyoxin D (Endorse ®)—a new fungicide for brown patch and large patch control. Purdue University
Pradeepkiran JA, Reddy AP, Reddy PH (2019) Pharmacophore-based models for therapeutic drugs against phosphorylated tau in Alzheimer’s disease. Drug Discov Today 24:616–623. https://doi.org/10.1016/J.DRUDIS.2018.11.005
Dixon SL, Smondyrev AM, Knoll EH et al (2006) PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. https://doi.org/10.1007/s10822-006-9087-6
An Y, Dong Y, Min L et al (2020) Construction and evaluation of molecular models: guide and design of novel SE inhibitors. ACS Publ 11:1152–1159. https://doi.org/10.1021/acsmedchemlett.0c00017
Sastry GM, Dixon SL, Sherman W (2011) Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J Chem Inf Model 51:2455–2466. https://doi.org/10.1021/ci2002704
Bowers KJ, Chow E, Xu H, et al (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06. ACM Press, New York, p 84
Liu X, Shi D, Zhou S et al (2017) Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov 13:23–37. https://doi.org/10.1080/17460441.2018.1403419
Bai Q, Liu S, Tian Y et al (2022) Application advances of deep learning methods for de novo drug design and molecular dynamics simulation. Wiley Interdiscip Rev Comput Mol Sci 12:e1581. https://doi.org/10.1002/WCMS.1581
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. https://doi.org/10.1371/journal.pone.0119264
Devi K, Patar L, Modi MK, Sen P (2017) An insight into structure, function, and expression analysis of 3-hydroxy-3-methylglutaryl-CoA reductase of Cymbopogon winterianus. Bioinform Biol Insights. https://doi.org/10.1177/1177932217701735
Wang E, Sun H, Wang J et al (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 119:9478–9508. https://doi.org/10.1021/ACS.CHEMREV.9B00055/ASSET/IMAGES/MEDIUM/CR-2019-000558_M019.GIF
Acknowledgements
All authors acknowledge the support provided by Department of Biochemistry and Forensic Science and the Department of Microbiology and Biotechnology (DST-FIST supported department). Author, P.R is thankful to ScHeme Of Developing High quality research (SHODH), Education department, Government of Gujarat, India for providing student support fellowship. Special thanks is extended to Dr. Rajendra Ku. Baharia, of ICMR-National Institute of Malaria Research (NIMR) Field Unit (FU), Nadiad, Gujarat for providing Aedes aegypti specie used for the experiments. All authors would like to thank Department of Chemistry and Department of Botany, Bioinformatics and Climate Change Impacts Management, School of Sciences at Gujarat University for allowing access to the advance instrumentation and the bioinformatics research facilities.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for profit sectors.
Author information
Authors and Affiliations
Contributions
PR and DG wrote the manuscript; PR, JN and MD performed experiments; PR prepared figures, and tables; RMR conceptualized the idea; RMR and DG critically proofread the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All authors have seen and approved the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rao, P., Ninama, J., Dudhat, M. et al. Curcumin interferes with chitin synthesis in Aedes aegypti: a computational and experimental investigation. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10672-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10672-0