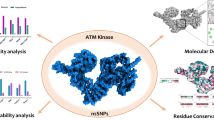
Abstract
The NEK6 (NIMA-related kinase 6) serine/threonine kinase is a pivotal player in a multitude of cellular processes, including the regulation of the cell cycle and the response to DNA damage. Its significance extends to disease pathogenesis, as changes in NEK6 activity have been linked to the development of cancer. Non-synonymous single nucleotide polymorphisms (nsSNPs) in NEK6 have been linked to cancer as they alter the protein’s native structure and function. The association between NEK6 activity and cancer development has prompted researchers to explore the effects of genetic variations within the NEK6 gene. Therefore, we utilized advanced computational tools to analyze 155 high-confidence nsSNPs in the NEK6 gene. From this analysis, 21 nsSNPs were identified as potentially harmful, raising concerns about their impact on NEK6 activity and cancer risk. These 21 mutations were then examined for structural alterations, and eight of nsSNPs (I51M, V76A, I134N, Y152D, R171Q, V186G, L237R, and C285S) were found to destabilize the protein. Among the destabilizing mutations screened, a specific mutation, R171Q, stood out due to its conserved nature. To understand its impact on the protein and conformation, all-atom molecular dynamics simulations (MDS) for 100 ns were performed for both Wildtype NEK6 (WT-NEK6) and R171Q. The simulations revealed that the R171Q variant was unstable and led to significant conformational changes in NEK6. This study provides valuable insights into NEK6 dysfunction caused by single amino acid alterations, offering a novel understanding of the molecular mechanisms underlying NEK6-related cancer progression.
Graphical abstract














Similar content being viewed by others
Data availability
Data will be shared upon reasonable request.
Consent to publish
Not applicable.
Abbreviations
- EVs:
-
Eigenvectors
- FATHMM:
-
Functional analysis through hidden Markov models
- FEL:
-
Free energy landscape
- HOPES:
-
High-throughput evaluation of protein stability
- NEK6:
-
NIMA (Never In Mitosis Gene A)-related kinase 6
- nsSNP:
-
Non-synonymous single nucleotide polymorphism
- MDS:
-
Molecular dynamics simulations
- PANTHER:
-
Protein ANalysis THrough Evolutionary Relationships
- PCA:
-
Principal component analysis
- PhD-SNP:
-
Predicting human deleterious single nucleotide polymorphisms
- PMut:
-
Protein mutation analysis
- PolyPhen-2:
-
Polymorphism Phenotyping v2
- Provean:
-
Protein variation effect analyzer
- Rg:
-
Radius of gyration
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- SASA:
-
Solvent accessible surface area
- SIFT:
-
Sorting intolerant from tolerant
- SNAP:
-
Screening for non-acceptable polymorphisms
- WT:
-
Wild-type
References
O’Regan L, Blot J, Fry AM (2007) Mitotic regulation by NIMA-related kinases. Cell Div 2:1–12. https://doi.org/10.1186/1747-1028-2-25
Panchal NK, Evan Prince S (2022) The NEK family of serine/threonine kinases as a biomarker for cancer. Clin Exp Med. https://doi.org/10.1007/s10238-021-00782-0
Sdelci S, Bertran MT, Roig J (2011) Nek9, Nek6, Nek7 and the separation of centrosomes. Cell Cycle 10:3816–3817. https://doi.org/10.4161/cc.10.22.18226
Li MZ, Yu L, Liu Q et al (1999) Assignment of NEK6, a NIMA-related gene, to human chromosome 9q33. 3 → q34.11 by radiation hybrid mapping. Cytogenet Cell Genet 87:271–272. https://doi.org/10.1159/000015445
Moraes EC, Meirelles GV, Honorato RV et al (2015) Kinase inhibitor profile for human nek1, nek6, and nek7 and analysis of the structural basis for inhibitor specificity. Molecules 20:1176–1191. https://doi.org/10.3390/molecules20011176
Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85:149–158. https://doi.org/10.1016/s0092-8674(00)81092-2
Belham C, Roig J, Caldwell JA et al (2003) A mitotic cascade of NIMA family kinases: Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem 278:34897–34909. https://doi.org/10.1074/jbc.M303663200
Meirelles GV, Silva JC, Mendonça YDA et al (2011) Human Nek6 is a monomeric mostly globular kinase with an unfolded short N-terminal domain. BMC Struct Biol. https://doi.org/10.1186/1472-6807-11-12
Atish DC, Anna CS, Maura BC et al (2017) Castration resistance in prostate cancer is mediated by the kinase NEK6. Cancer Res 77:753–765. https://doi.org/10.1158/0008-5472.CAN-16-0455
Jee HJ, Kim AJ, Song N et al (2010) Nek6 overexpression antagonizes p53-induced senescence in human cancer cells. Cell Cycle 9:4703–4710. https://doi.org/10.4161/cc.9.23.14059
De Donato M, Fanelli M, Mariani M et al (2015) Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Am J Cancer Res 5:1862–1877
Cao F, Wu X, Shan Y et al (2021) Circular RNA NEK6 contributes to the development of non-small-cell lung cancer by competitively binding with miR-382-5p to elevate BCAS2 expression at post-transcriptional level. BMC Pulm Med 21:325. https://doi.org/10.1186/s12890-021-01617-0
Jee HJ, Kim H-J, Kim AJ et al (2011) Nek6 suppresses the premature senescence of human cancer cells induced by camptothecin and doxorubicin treatment. Biochem Biophys Res Commun 408:669–673. https://doi.org/10.1016/j.bbrc.2011.04.083
Sampson J, O’Regan L, Dyer MJS et al (2017) Hsp72 and Nek6 cooperate to cluster amplified centrosomes in cancer cells. Cancer Res 77:4785–4796. https://doi.org/10.1158/0008-5472.CAN-16-3233
De DM, Fanelli M, Mariani M et al (2015) Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Am J Cancer Res 5:1862–1877
Choudhury AD, Schinzel AC, Cotter MB et al (2017) Castration resistance in prostate cancer is mediated by the kinase NEK6. Cancer Res 77:753–765. https://doi.org/10.1158/0008-5472.CAN-16-0455
Desai M, Chauhan JB (2018) Computational analysis for the determination of deleterious nsSNPs in human MTHFR gene. Comput Biol Chem 74:20–30. https://doi.org/10.1016/j.compbiolchem.2018.02.022
Desai M, Chauhan JB (2017) Computational analysis for the determination of deleterious nsSNPs in human MTHFD1 gene. Comput Biol Chem 70:7–14. https://doi.org/10.1016/j.compbiolchem.2017.07.001
Dash R, Munni YA (2020) Computational SNP analysis and molecular simulation revealed the most computational SNP analysis and molecular simulation revealed the most deleterious missense variants in the NBD1 domain of human ABCA1 transporter. https://doi.org/10.3390/ijms21207606
Solayman M, Saleh MA, Paul S et al (2017) In silico analysis of nonsynonymous single nucleotide polymorphisms of the human adiponectin receptor 2 (ADIPOR2) gene. Comput Biol Chem 68:175–185. https://doi.org/10.1016/j.compbiolchem.2017.03.005
Panchal NK, Bhale A, Verma VK, Beevi SS (2020) Computational and molecular dynamics simulation approach to analyze the impact of XPD gene mutation on protein stability and function. Mol Simul 46:1200–1219. https://doi.org/10.1080/08927022.2020.1810852
Tanwar G, Mazumder AG, Bhardwaj V et al (2019) Target identification, screening and in vivo evaluation of pyrrolone-fused benzosuberene compounds against human epilepsy using Zebrafish model of pentylenetetrazol-induced seizures. Sci Rep. https://doi.org/10.1038/s41598-019-44264-6
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2012) In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ). FEBS Open Bio 2:285–293. https://doi.org/10.1016/j.fob.2012.09.003
Bhardwaj V, Purohit R (2020) Computational investigation on effect of mutations in PCNA resulting in structural perturbations and inhibition of mismatch repair pathway. J Biomol Struct Dyn 38:1963–1974. https://doi.org/10.1080/07391102.2019.1621210
Kumar Bhardwaj V, Purohit R, Kumar S (2021) Himalayan bioactive molecules as potential entry inhibitors for the human immunodeficiency virus. Food Chem. https://doi.org/10.1016/j.foodchem.2020.128932
Jee HJ, Kim H-J, Kim AJ et al (2013) The inhibition of Nek6 function sensitizes human cancer cells to premature senescence upon serum reduction or anticancer drug treatment. Cancer Lett 335:175–182. https://doi.org/10.1016/j.canlet.2013.02.012
O’Regan L, Fry AM (2009) The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol Cell Biol 29:3975–3990. https://doi.org/10.1128/mcb.01867-08
Mottaz A, David FPA, Veuthey AL, Yip YL (2010) Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics 26:851–852. https://doi.org/10.1093/bioinformatics/btq028
López-Ferrando V, Gazzo A, De La Cruz X et al (2017) PMut: a web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res 45:W222–W228. https://doi.org/10.1093/nar/gkx313
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. https://doi.org/10.1093/bioinformatics/btv195
Savojardo C, Fariselli P, Martelli PL, Casadio R (2016) INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics 32:2542–2544. https://doi.org/10.1093/bioinformatics/btw192
Worth CL, Preissner R, Blundell TL (2011) SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 39:215–222. https://doi.org/10.1093/nar/gkr363
Pires DEV, Ascher DB, Blundell TL (2014) MCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30:335–342. https://doi.org/10.1093/bioinformatics/btt691
Pires DEV, Ascher DB, Blundell TL (2014) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 42:314–319. https://doi.org/10.1093/nar/gku411
Parthiban V, Gromiha MM, Schomburg D (2006) CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res 34:239–242. https://doi.org/10.1093/nar/gkl190
Rogers MF, Shihab HA, Mort M et al (2018) FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34:511–513. https://doi.org/10.1093/bioinformatics/btx536
Celniker G, Nimrod G, Ashkenazy H et al (2013) ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr J Chem 53:199–206. https://doi.org/10.1002/ijch.201200096
Ashkenazy H, Abadi S, Martz E et al (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350. https://doi.org/10.1093/nar/gkw408
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. https://doi.org/10.1038/nprot.2010.5
Parra RG, Schafer NP, Radusky LG et al (2016) Protein Frustratometer 2: a tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res 44:W356–W360. https://doi.org/10.1093/NAR/GKW304
Seeliger D, De Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24:417–422. https://doi.org/10.1007/s10822-010-9352-6
Panchal NK, Mohanty S, Prince SE (2023) NIMA-related kinase-6 (NEK6) as an executable target in cancer. Clin Transl Oncol 25:66–77
Jessica Montgomery BM (2016) Nek6 controls mitotic progression through regulating Eml3 localisation to spindle microtubules. Thesis
Yin M-J, Shao L, Voehringer D et al (2003) The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J Biol Chem 278:52454–52460. https://doi.org/10.1074/jbc.M308080200
Lee M-Y, Kim H-J, Kim M-A et al (2008) Nek6 is involved in G2/M phase cell cycle arrest through DNA damage-induced phosphorylation. Cell Cycle 7:2705–2709. https://doi.org/10.4161/cc.7.17.6551
Acknowledgements
All the authors are grateful to the Vellore Institute of Technology, Vellore, India for providing the essential facilities to carry out this work.
Funding
No funding has been received for this work.
Author information
Authors and Affiliations
Contributions
" N.K.P and S.E.P: Conceptualization, Literature survey; N.K.P and S.M : Performed experiment ; N.K.P: Wrote the Original draft ; S.E.P: Supervised the work. All authors reviewed the manuscript "
Corresponding author
Ethics declarations
Conflict of interest
We affirm that there are no conflicts of interest to disclose regarding the publication of this manuscript.
Ethical approval
Not applicable.
Informed consent
Not applicable: no human/animal subjects were involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Panchal, N.K., Mohanty, S. & Prince, S.E. Computational insights into NIMA-related kinase 6: unraveling mutational effects on structure and function. Mol Cell Biochem (2023). https://doi.org/10.1007/s11010-023-04910-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11010-023-04910-0