Abstract
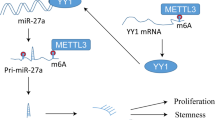
Methyltransferase like 3 (METTL3) has been reported to promote tumorigenesis of multiple myeloma (MM), however, the molecular mechanism still needs further research. The N6-methyladenosine (m6A) level in tissues or cells was measured by m6A kit and dot blot assay. The mRNA and protein expression were detected by quantitative real-time PCR (RT-qPCR) and Western blot, respectively. The cell counting kit-8 and colony formation assay were used to detect the cell proliferation. Coimmunoprecipitation (Co-IP) experiment verified the binding of two proteins. The luciferase reporter experiment demonstrated the targeted binding of miR-182-5p and CaMKII inhibitor 1 (CAMK2N1). More importantly, tumor growth was measured in xenograft mice. Our data showed that the expression of METTL3 was significantly increased in MM patients’ samples and MM cells. METTL3 overexpression promoted MM cells proliferation, and METTL3 knockdown inhibited MM cells proliferation. Mechanically, METTL3-dependent m6A participated in DiGeorge syndrome critical region 8 (DGCR8)-mediated maturation of pri-miR-182. Upregulation of miR-182-5p further enhanced the promoting proliferation effect of METTL3 overexpression on MM cells. Moreover, the luciferase reporter gene experiment proved that miR-182-5p targetedly regulated CAMK2N1 expression. Xenograft tumor in nude mice further verified that METTL3 promoted MM tumor growth through miR-182/CAMK2N1 signal axis. In summary, the METTL3/miR-182-5p/CAMK2N1 axis plays an important role in MM tumorigenesis, which may provide a new target for MM therapy.








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, Tuazon S, Gopal AK, Libby EN (2022) Diagnosis and management of multiple myeloma: a review. JAMA 327:464–477. https://doi.org/10.1001/jama.2022.0003
Liu J, Liu W, Mi L, Zeng X, Cai C, Ma J, Wang L (2019) Incidence and mortality of multiple myeloma in China, 2006–2016: an analysis of the Global Burden of Disease Study 2016. J Hematol Oncol 12:136. https://doi.org/10.1186/s13045-019-0807-5
Zhao WH, Wang BY, Chen LJ, Fu WJ, Xu J, Liu J, Jin SW, Chen YX, Cao XM, Yang Y, Zhang YL, Wang FX, Zhang PY, Lei B, Gu LF, Wang JL, Zhang H, Bai J, Xu Y, Zhu H, Du J, Jiang H, Fan XH, Li JY, Hou J, Chen Z, Zhang WG, Mi JQ, Chen SJ, He AL (2022) Four-year follow-up of LCAR-B38M in relapsed or refractory multiple myeloma: a phase 1, single-arm, open-label, multicenter study in China (LEGEND-2). J Hematol Oncol 15:86. https://doi.org/10.1186/s13045-022-01301-8
Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, Zhang R (2020) Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res 48:6251–6264. https://doi.org/10.1093/nar/gkaa347
Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, Wei Z, Su J, Liu G, Rong R, Lu Z, de Magalhães JP, Rigden DJ, Meng J (2021) m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res 49:D134-d143. https://doi.org/10.1093/nar/gkaa692
Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, Zhao J, Gao S, Jurczyszyn A, Janz S, Beksac M, Zhan F, Gu C, Yang Y (2021) HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J Hematol Oncol 14:54. https://doi.org/10.1186/s13045-021-01066-6
Song S, Fan G, Li Q, Su Q, Zhang X, Xue X, Wang Z, Qian C, Jin Z, Li B (2021) IDH2 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in multiple myeloma. Oncogene 40:5393–5402. https://doi.org/10.1038/s41388-021-01939-7
Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao F, Fan F, Xu J, Zhang B, Zhang Y, Yin X, Cheng Q, Gao S, Deng J, Mei H, Huang Z, Sun C, Hu Y (2022) FTO promotes multiple myeloma progression by posttranscriptional activation of HSF1 in an m(6)A-YTHDF2-dependent manner. Mol Ther 30:1104–1118. https://doi.org/10.1016/j.ymthe.2021.12.012
Che F, Ye X, Wang Y, Wang X, Ma S, Tan Y, Mao Y, Luo Z (2022) METTL3 facilitates multiple myeloma tumorigenesis by enhancing YY1 stability and pri-microRNA-27 maturation in m(6)A-dependent manner. Cell Biol Toxicol. https://doi.org/10.1007/s10565-021-09690-1
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M (2016) m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540:301–304. https://doi.org/10.1038/nature20577
Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vågbø CB, Kusśnierczyk A, Klungland A, Darnell JE Jr, Darnell RB (2015) A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29:2037–2053. https://doi.org/10.1101/gad.269415.115
Lichinchi G, Gao S, Saletore Y, Gonzalez GM (2016) Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. https://doi.org/10.1038/nmicrobiol.2016.11
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. https://doi.org/10.1038/nature12730
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015) N6-methyladenosine marks primary microRNAs for processing. Nature 519:482–485. https://doi.org/10.1038/nature14281
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF (2015) HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162:1299–1308. https://doi.org/10.1016/j.cell.2015.08.011
Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC (2017) Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep 20:2262–2276. https://doi.org/10.1016/j.celrep.2017.08.027
Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, Xi J, Ye D, Zhu S, Chen W, Jia W, Leng Y, Wan X, Kang J (2018) N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res 46:3906–3920. https://doi.org/10.1093/nar/gky130
Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY, Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, Li KS, Gao DM, Ma DN, Ye BG, Wang CH, Qin CD, Sun HC, Zhang T, Tang ZY (2018) miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol 11:12. https://doi.org/10.1186/s13045-018-0555-y
Wu X, Wang W (2021) miR-182–5p serves as an oncogene in lung adenocarcinoma through binding to STARD13. Comput Math Meth Med 2021:7074343. https://doi.org/10.1155/2021/7074343
Souza MF, Cólus IMS (2022) MiR-182–5p modulates prostate cancer aggressive phenotypes by targeting EMT associated pathways. Biomolecules. 12:187. https://doi.org/10.3390/biom12020187
Du K, Zhang Z, Zeng Z, Tang J, Lee T, Sun T (2021) Distinct roles of Fto and Mettl3 in controlling development of the cerebral cortex through transcriptional and translational regulations. Cell Death Dis 12:700. https://doi.org/10.1038/s41419-021-03992-2
Wang D, Wang X, Huang B, Zhao Y, Tu W, Jin X, Shao Y, Zhu Y, Lu G (2022) METTL3 promotes prostate cancer progression by regulating miR-182 maturation in m6A-dependent manner. Andrologia 54:1581–1591. https://doi.org/10.1111/and.14422
Zhang X, Tian L, Li Z, Liu R, Yu J, Liu B (2022) CAMK2N1 has a cancer-suppressive function in colorectal carcinoma via effects on the Wnt/β-catenin pathway. Biochem Biophys Res Commun 626:220–228. https://doi.org/10.1016/j.bbrc.2022.08.036
Xu K, Hu X, Sun L, Liang Q, Ouyang G, Zhang Y, Mu Q, Yan X (2019) MicroRNA-532 exerts oncogenic functions in t(4;14) multiple myeloma by targeting CAMK2N1. Hum Cell 32:529–539. https://doi.org/10.1007/s13577-019-00276-y
Wang J, Zuo Y, Lv C, Zhou M, Wan Y (2023) N6-methyladenosine regulators are potential prognostic biomarkers for multiple myeloma. IUBMB Life 75:137–148. https://doi.org/10.1002/iub.2678
Zeng C, Huang W, Li Y, Weng H (2020) Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol 13:117. https://doi.org/10.1186/s13045-020-00951-w
Huang X, Yang Z, Li Y, Long X (2023) m6A methyltransferase METTL3 facilitates multiple myeloma cell growth through the m6A modification of BZW2. Ann Hematol 102:1801–1810. https://doi.org/10.1007/s00277-023-05283-6
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, Wei JF, Yang H (2019) METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer 18:110. https://doi.org/10.1186/s12943-019-1036-9
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y (2019) Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res 38:393. https://doi.org/10.1186/s13046-019-1408-4
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y (2021) METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther 28:335–349. https://doi.org/10.1038/s41417-020-00222-3
Yao B, Zhu S, Wei X, Chen MK, Feng Y, Li Z, Xu X, Zhang Y, Wang Y, Zhou J, Tang N, Ji C, Jiang P, Zhao SC, Qin C, Feng N (2022) The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol Cancer 21:119. https://doi.org/10.1186/s12943-022-01598-6
Acknowledgements
Not applicable.
Funding
This research was supported by Anhui Provincial Department of Education (2022AH051178), the Anhui Natural Science Foundation of China (2008085MH296), the Anhui Medical University Natural Science Foundation (2020xkj186).
Author information
Authors and Affiliations
Contributions
JB, TTX, WJW, HX and XWC collected clinical data. JB and TTX carried out the experiments and analyzed this data. The manuscript was written and interpreted by JB. Study design, experiments and manuscript review were handled by RXX. The final manuscript was approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
For patients: This project was approved by the Research Ethics Committee of Anhui Medical University (No. 20200040). All participants in this project signed the informed consent form.
For animals: The animal experiments were carried out with the approval of the ethics committee of Anhui Medical University (No. 20190657).
Consent for publication
Not applicable.
Competing interests
All authors declare no competing interests.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bao, J., Xu, T., Wang, W. et al. N6-methyladenosine-induced miR-182-5p promotes multiple myeloma tumorigenesis by regulating CAMK2N1. Mol Cell Biochem (2024). https://doi.org/10.1007/s11010-023-04906-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11010-023-04906-w