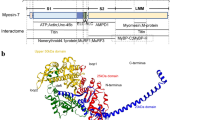
Abstract
Duchenne Muscular Dystrophy (DMD) is a progressive muscle wasting disorder caused by loss-of-function mutations in the dystrophin gene. Although the search for a definitive cure has failed to date, extensive efforts have been made to introduce effective therapeutic strategies. Gene editing technology is a great revolution in biology, having an immediate application in the generation of research models. DMD muscle cell lines are reliable sources to evaluate and optimize therapeutic strategies, in-depth study of DMD pathology, and screening the effective drugs. However, only a few immortalized muscle cell lines with DMD mutations are available. In addition, obtaining muscle cells from patients also requires an invasive muscle biopsy. Mostly DMD variants are rare, making it challenging to identify a patient with a particular mutation for a muscle biopsy. To overcome these challenges and generate myoblast cultures, we optimized a CRISPR/Cas9 gene editing approach to model the most common DMD mutations that include approximately 28.2% of patients. GAP-PCR and sequencing results show the ability of the CRISPR-Cas9 system to efficient deletion of mentioned exons. We showed producing truncated transcript due to the targeted deletion by RT-PCR and sequencing. Finally, mutation-induced disruption of dystrophin protein expression was confirmed by western blotting. All together, we successfully created four immortalized DMD muscle cell lines and showed the efficacy of the CRISPR-Cas9 system for the generation of immortalized DMD cell models with the targeted deletions.
Graphical abstract






Similar content being viewed by others
Data availability
Enquiries about data availability should be directed to the authors.
References
Mercuri E, Muntoni F (2013) Muscular dystrophies. Lancet 381(9869):845–860
Amoasii L, Long C, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E et al (2017) Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 9(418):8081
Shimizu-Motohashi Y, Miyatake S, Komaki H, Si T, Aoki Y (2016) Recent advances in innovative therapeutic approaches for Duchenne muscular dystrophy: from discovery to clinical trials. Am J Transl Res 8(6):2471
Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L et al (2010) Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol 9(2):177–189
Emery AEH (2002) The muscular dystrophies. Lancet 359(9307):687–695
Duchêne B, Iyombe-Engembe JP, Rousseau J, Tremblay JP, Ouellet DL (2018) From gRNA Identification to the restoration of dystrophin expression: A dystrophin gene correction strategy for duchenne muscular dystrophy mutations using the crispr-induced deletion method. In: Bernardini C (ed) duchenne muscular dystrophy. Springer, New York, pp 267–83
Ellis JA, Vroom E, Muntoni F (2013) 195th ENMC international workshop: newborn screening for Duchenne muscular dystrophy 14–16th December, 2012, Naarden. Netherlands Neuromuscul Dis 23(8):682–689
Nguyen Q, Yokota T (2017) Immortalized muscle cell model to test the exon skipping efficacy for Duchenne muscular dystrophy. J Pers Med 7(4):13
Lim KRQ, Nguyen Q, Dzierlega K, Huang Y, Yokota T (2020) CRISPR-generated animal models of duchenne muscular dystrophy. Genes 11(3):342
Wasala NB, Chen SJ, Duan D (2020) Duchenne muscular dystrophy animal models for high-throughput drug discovery and precision medicine. Expert Opin Drug Discov 15(4):443–456
Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM (1987) Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50(3):509–517
Egorova TV, Zotova ED, Reshetov DA, Polikarpova AV, Vassilieva SG, Vlodavets DV et al (2019) CRISPR/Cas9-generated mouse model of Duchenne muscular dystrophy recapitulating a newly identified large 430 kb deletion in the human DMD gene. Dis Models Mech 12(4):dmm037655
Gao Q, McNally EM (2015) The Dystrophin complex: structure, function and implications for therapy. Compr Physiol 5(3):1223
Ervasti JM (2007) Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta (BBA)-Mol Basis Dis 1772(2):108–117
Lapidos KA, Kakkar R, McNally EM (2004) The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res 94(8):1023–1031
Rader EP, Turk R, Willer T, Beltrán D, Inamori KI, Peterson TA et al (2016) Role of dystroglycan in limiting contraction-induced injury to the sarcomeric cytoskeleton of mature skeletal muscle. Proc Natl Acad Sci USA 113(39):10992–10997
Pearce JMS, Pennington RJT, Walton JN (1964) Serum enzyme studies in muscle disease: Part III Serum creatine kinase activity in relatives of patients with the Duchenne type of muscular dystrophy. J Neurol Neurosurg Psychiatry 27(3):181
Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S et al (2016) A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 18(4):533–540
Yeung EW, Whitehead NP, Suchyna TM, Gottlieb PA, Sachs F, Allen DG (2005) Effects of stretch-activated channel blockers on [Ca2+] i and muscle damage in the mdx mouse. J Physiol 562(2):367–380
Dowling P, Doran P, Ohlendieck K (2004) Drastic reduction of sarcalumenin in Dp427 (dystrophin of 427 kDa)-deficient fibres indicates that abnormal calcium handling plays a key role in muscular dystrophy. Biochem J 379(2):479–488
Nghiem PP, Kornegay JN (2019) Gene therapies in canine models for Duchenne muscular dystrophy. Hum Genet 138(5):483–489
Nakamura A, Si T (2011) Mammalian models of Duchenne muscular dystrophy: pathological characteristics and therapeutic applications. J Biomed Biotechnol. https://doi.org/10.1155/2011/184393
Pane M, Scalise R, Berardinelli A, D’Angelo G, Ricotti V, Alfieri P et al (2013) Early neurodevelopmental assessment in Duchenne muscular dystrophy. Neuromuscul Disord 23(6):451–455
Jelinkova S, Markova L, Pesl M, Valášková I, Makaturová E, Jurikova L et al (2019) Generation of two Duchenne muscular dystrophy patient-specific induced pluripotent stem cell lines DMD02 and DMD03 (MUNIi001-A and MUNIi003-A). Stem Cell Res 40:101562
Nguyen Q, Yokota T (2019) Antisense oligonucleotides for the treatment of cardiomyopathy in Duchenne muscular dystrophy. Am J Transl Res 11(3):1202
Nachman MW (2004) Haldane and the first estimates of the human mutation rate. J Genet 83(3):231–233
Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K et al (2015) The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat 36(4):395–402
Yavas A, Weij R, van Putten M, Kourkouta E, Beekman C, Puoliväli J et al (2020) Detailed genetic and functional analysis of the hDMDdel52/mdx mouse model. PLoS ONE 15(12):e0244215
Echigoya Y, Lim KRQ, Nakamura A, Yokota T (2018) Multiple exon skipping in the Duchenne muscular dystrophy hot spots: prospects and challenges. J Pers Med 8(4):41
Shimo T, Hosoki K, Nakatsuji Y, Yokota T, Obika S (2018) A novel human muscle cell model of Duchenne muscular dystrophy created by CRISPR/Cas9 and evaluation of antisense-mediated exon skipping. J Hum Genet 63(3):365–375
Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2(1):90–95
Duchêne BL, Cherif K, Iyombe-Engembe JP, Guyon A, Rousseau J, Ouellet DL et al (2018) CRISPR-induced deletion with SaCas9 restores dystrophin expression in dystrophic models in vitro and in vivo. Mol Ther 26(11):2604–2616
Aartsma-Rus A, Van Deutekom JCT, Fokkema IF, Van Ommen GJB, Den Dunnen JT (2006) Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve Off J Am Assoc Electrodiagn Med 34(2):135–144
Min YL, Chemello F, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Mireault AA et al (2020) Correction of three prominent mutations in mouse and human models of Duchenne muscular dystrophy by single-cut genome editing. Mol Ther 28(9):2044–2055
McGreevy JW, Hakim CH, McIntosh MA, Duan D (2015) Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech 8(3):195–213
Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T et al (2015) Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep 4(1):143–154
Iyombe-Engembe JP, Ouellet DL, Barbeau X, Rousseau J, Chapdelaine P, Lagüe P et al (2016) Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol Ther Nucleic Acids 5:e283
Mendell JR, Rodino-Klapac L, Sahenk Z, Malik V, Kaspar BK, Walker CM et al (2012) Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci Lett 527(2):90–99
DaM N, Lindsay A, Judge LM, Duan D, Chamberlain JS, Lowe DA et al (2018) Variable rescue of microtubule and physiological phenotypes in mdx muscle expressing different miniaturized dystrophins. Hum Mol Genet 27(12):2090–100
Le Guiner C, Servais L, Montus M, Larcher T, Fraysse B, Moullec S et al (2017) Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat Commun 8(1):1–15
Le Hir M, Goyenvalle A, Peccate C, Précigout G, Davies KE, Voit T et al (2013) AAV genome loss from dystrophic mouse muscles during AAV-U7 snRNA-mediated exon-skipping therapy. Mol Ther 21(8):1551–1558
Xiang X, Zhao X, Pan X, Dong Z, Yu J, Li S et al (2021) Efficient correction of Duchenne muscular dystrophy mutations by SpCas9 and dual gRNAs. Mol Ther Nucleic Acids 24:403–415
Sun C, Shen L, Zhang Z, Xie X (2020) Therapeutic strategies for Duchenne muscular dystrophy: an update. Genes 11(8):837
Karkare S, Bhatnagar D (2006) Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl Microbiol Biotechnol 71(5):575–586
Niks EH, Aartsma-Rus A (2017) Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin Biol Ther 17(2):225–236
Aartsma-Rus A, Van Ommen GJB (2007) Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. Rna 13(10):1609–24
Young CS, Mokhonova E, Quinonez M, Pyle AD, Spencer MJ (2017) Creation of a novel humanized dystrophic mouse model of duchenne muscular dystrophy and application of a CRISPR/Cas9 gene editing therapy. J Neuromuscul Dis 4(2):139–145
Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA (2015) Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 6(1):1–13
Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN (2014) Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science 345(6201):1184–1188
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Ousterout DG, Perez-Pinera P, Thakore PI, Kabadi AM, Brown MT, Qin X et al (2013) Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther 21(9):1718–1726
Popplewell L, Koo T, Leclerc X, Duclert A, Mamchaoui K, Gouble A et al (2013) Gene correction of a duchenne muscular dystrophy mutation by meganuclease-enhanced exon knock-in. Hum Gene Ther 24(7):692–701
Rousseau J, Chapdelaine P, Boisvert S, Almeida LP, Corbeil J, Montpetit A et al (2011) Endonucleases: tools to correct the dystrophin gene. J Gene Med 13(10):522–537
Min YL, Bassel-Duby R, Olson EN (2019) CRISPR correction of Duchenne muscular dystrophy. Annu Rev Med 70:239
Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H et al (2018) Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 4(1):eaap9004
Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM et al (2019) CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 5(3):eaav4324
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ et al (2016) In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540(7631):144–149
Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST et al (2018) Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36(6):536–539
Eslahi A, Alizadeh F, Avan A, Ferns GA, Moghbeli M, Abbaszadegan MR et al (2023) New advancements in CRISPR based gene therapy of Duchenne muscular dystrophy. Gene. https://doi.org/10.1016/j.gene.2023.147358
Cai M, Yang Y (2014) Targeted genome editing tools for disease modeling and gene therapy. Curr Gene Ther 14(1):2–9
Lu XJ, Ji LJ, Kato T, Takada S (2017) In vivo and in vitro disease modeling with CRISPR/Cas9. Brief Funct Genomics 16(1):13–24
Takizawa H, Hara Y, Mizobe Y, Ohno T, Suzuki S, Inoue K et al (2019) Modelling Duchenne muscular dystrophy in MYOD1-converted urine-derived cells treated with 3-deazaneplanocin A hydrochloride. Sci Rep 9(1):1–11
Maruyama R, Yokota T (2018) Creation of DMD muscle cell model using CRISPR-Cas9 genome editing to test the efficacy of antisense-mediated exon skipping. In: Yokota T, Maruyama R (eds) Exon Skipping and Inclusion Therapies. Springer, New York, pp 165–171
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93(3):1156–1160
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A et al (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186(2):757–761
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE et al (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Wang H, La Russa M, Qi LS (2016) CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 85(1):227–264
Martinez-Lage M, Torres-Ruiz R, Rodriguez-Perales S (2017) CRISPR/Cas9 technology: applications and human disease modeling. Prog Mol Biol Transl Sci 152:23–48
Heidenreich M, Zhang F (2016) Applications of CRISPR–Cas systems in neuroscience. Nat Rev Neurosci 17(1):36–44
Torres-Ruiz R, Rodriguez-Perales S (2015) CRISPR-Cas9: a revolutionary tool for cancer modelling. Int J Mol Sci 16(9):22151–22168
Seeger T, Porteus M, Wu JC (2017) Genome editing in cardiovascular biology. Circ Res 120(5):778–780
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308
Mamchaoui K, Trollet C, Bigot A, Negroni E, Chaouch S, Wolff A et al (2011) Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet Muscle 1(1):1–11
Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, Van Deutekom J, van Ommen GJ et al (2009) Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat 30(3):293–299
Renault V, Thorne LE, Eriksson PO, Butler-Browne G, Mouly V (2002) Regenerative potential of human skeletal muscle during aging. Aging Cell 1(2):132–139
Wright WE, Shay JW (2002) Historical claims and current interpretations of replicative aging. Nat Biotechnol 20(7):682–688
Webster C, Blau HM (1990) Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat Cell Mol Genet 16(6):557–565
Périé S, Mamchaoui K, Mouly V, Blot S, Bouazza B, Thornell L-E et al (2006) Premature proliferative arrest of cricopharyngeal myoblasts in oculo-pharyngeal muscular dystrophy: therapeutic perspectives of autologous myoblast transplantation. Neuromuscul Disord 16(11):770–781
Aartsma-Rus A, Bremmer-Bout M, Janson AAM, den Dunnen JT, van Ommen GJB, van Deutekom JCT (2002) Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul Disord 12:S71–S7
Aartsma-Rus A, De Winter CL, Janson AAM, Kaman WE, Van Ommen GJB, Den Dunnen JT et al (2005) Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides 15(4):284–197
Aartsma-Rus A, Janson AAM, van Ommen GJB, van Deutekom JCT (2007) Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med Genet 8(1):1–9
Popplewell LJ, Adkin C, Arechavala-Gomeza V, Aartsma-Rus A, de Winter CL, Wilton SD et al (2010) Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscul Disord 20(2):102–110
Echigoya Y, Lim KRQ, Trieu N, Bao B, Nichols BM, Vila MC et al (2017) Quantitative antisense screening and optimization for exon 51 skipping in Duchenne muscular dystrophy. Mol Ther 25(11):2561–2572
Echigoya Y, Mouly V, Garcia L, Yokota T, Duddy W (2015) In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS ONE 10(3):e0120058
Chaouch S, Mouly V, Goyenvalle A, Vulin A, Mamchaoui K, Negroni E et al (2009) Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther 20(7):784–790
Lu XJ, Ji LJ, Torres-Ruiz R, Rodriguez-Perales S (2017) CRISPR-Cas9 technology: applications and human disease modelling. Brief Funct Genomics 16(1):4–12
Bauer DE, Canver MC, Orkin SH (2015) Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. JoVE (J Vis Exp) 95:e52118
Brescia M, Janssen JM, Liu J, Gonçalves MAFV (2020) High-capacity adenoviral vectors permit robust and versatile testing of DMD gene repair tools and strategies in human cells. Cells 9(4):869
Maggio I, Chen X, Gonçalves MAFV (2016) The emerging role of viral vectors as vehicles for DMD gene editing. Genome Med 8(1):1–10
Maggio I, Liu J, Janssen JM, Chen X, Gonçalves MAFV (2016) Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep 6(1):1–12
Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z et al (2015) Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol 33(2):175–178
Kim D, Bae S, Park J, Kim E, Kim S, Yu HR et al (2015) Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12(3):237–243
Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK (2017) CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat Methods 14(6):607–614
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE et al (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6):1380–1389
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32(3):279–284
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D et al (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32(6):569–576
Guilinger JP, Thompson DB, Liu DR (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32(6):577–582
Tycko J, Myer VE, Hsu PD (2016) Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell 63(3):355–370
Kim S, Kim D, Cho SW, Kim J, Kim J-S (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24(6):1012–1019
Ramakrishna S, Dad ABK, Beloor J, Gopalappa R, Lee SK, Kim H (2014) Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24(6):1020–1027
Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O et al (2018) Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563(7733):646–651
Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P et al (2019) Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol 37(1):64–72
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.M.; methodology, F.A., Y.J and Y.M.; investigation, F.A and Y.J; writing—original draft preparation, F.A.; discussed and commented on the manuscript, A.E, SH.F and Y.Y; editing article, M.M.; visualization, F.A; supervision, M.M.; project administration, A.E.; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alizadeh, F., Abraghan, Y.J., Farrokhi, S. et al. Production of Duchenne muscular dystrophy cellular model using CRISPR-Cas9 exon deletion strategy. Mol Cell Biochem (2023). https://doi.org/10.1007/s11010-023-04759-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11010-023-04759-3