Abstract
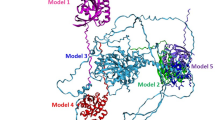
Quinine, a bitter compound, can act as an agonist to activate the family of bitter taste G protein-coupled receptor family of proteins. Previous work from our laboratory has demonstrated that quinine causes activation of RalA, a Ras p21-related small G protein. Ral proteins can be activated directly or indirectly through an alternative pathway that requires Ras p21 activation resulting in the recruitment of RalGDS, a guanine nucleotide exchange factor for Ral. Using normal mammary epithelial (MCF-10A) and non-invasive mammary epithelial (MCF-7) cell lines, we investigated the effect of quinine in regulating Ras p21 and RalA activity. Results showed that in the presence of quinine, Ras p21 is activated in both MCF-10A and MCF-7 cells; however, RalA was inhibited in MCF-10A cells, and no effect was observed in the case of MCF-7 cells. MAP kinase, a downstream effector for Ras p21, was activated in both MCF-10A and MCF-7 cells. Western blot analysis confirmed the expression of RalGDS in MCF-10A cells and MCF-7 cells. The expression of RalGDS was higher in MCF-10A cells in comparison to the MCF-7 cells. Although RalGDS was detected in MCF-10A and MCF-7 cells, it did not result in RalA activation upon Ras p21 activation with quinine suggesting that the Ras p21-RalGDS-RalA pathway is not active in the MCF-10A cells. The inhibition of RalA activity in MCF-10A cells due to quinine could be as a result of a direct effect of this bitter compound on RalA. Protein modeling and ligand docking analysis demonstrated that quinine can interact with RalA through the R79 amino acid, which is located in the switch II region loop of the RalA protein. It is possible that quinine causes a conformational change that results in the inhibition of RalA activation even though RalGDS is present in the cell. More studies are needed to elucidate the mechanism(s) that regulate Ral activity in mammary epithelial cells.








Similar content being viewed by others
References
Wheeler-Jones CP (2005) Cell signalling in the cardiovascular system: an overview. Heart 91(10):1366–1374
McCormick F (2019) Progress in targeting RAS with small molecule drugs. Biochem J 476(2):365–374
Chang EH et al (1982) Human genome contains four genes homologous to transforming genes of Harvey and Kirsten murine sarcoma viruses. Proc Natl Acad Sci USA 79(16):4848–4852
Tsuchida N, Murugan AK, Grieco M (2016) Kirsten Ras* oncogene: significance of its discovery in human cancer research. Oncotarget 7(29):46717–46733
Karnoub AE, Weinberg RA (2008) Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 9(7):517–531
Schubbert S, Shannon K, Bollag G (2007) Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 7(4):295–308
Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3(1):11–22
Wennerberg K, Rossman KL, Der CJ (2005) The Ras superfamily at a glance. J Cell Sci 118(Pt 5):843–846
Buday L, Vas V (2020) Novel regulation of Ras proteins by direct tyrosine phosphorylation and dephosphorylation. Cancer Metastasis Rev 39(4):1067–1073
Veleri S et al (2018) Molecular insights into the roles of Rab proteins in intracellular dynamics and neurodegenerative diseases. Neuromolecular Med 20(1):18–36
Clark GJ, Der CJ (1995) Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast Cancer Res Treat 35(1):133–144
Shields JM et al (2000) Understanding Ras: ’it ain’t over “til it’s over.” Trends Cell Biol 10(4):147–154
Keeton AB, Salter EA, Piazza GA (2017) The RAS-effector interaction as a drug target. Cancer Res 77(2):221–226
Hibino K et al (2011) Activation kinetics of RAF protein in the ternary complex of RAF, RAS-GTP, and kinase on the plasma membrane of living cells: single-molecule imaging analysis. J Biol Chem 286(42):36460–36468
Khanal P et al (2010) The prolyl isomerase Pin1 enhances HER-2 expression and cellular transformation via its interaction with mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1. Mol Cancer Ther 9(3):606–616
Santarpia L, Lippman SM, El-Naggar AK (2012) Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 16(1):103–119
Tian H et al (2018) Raf1 is a prognostic factor for progression in patients with nonsmall cell lung cancer after radiotherapy. Oncol Rep 39(4):1966–1974
Li S et al (2018) Inhibition of Raf1 ameliorates bleomycin-induced pulmonary fibrosis through attenuation of TGF-beta1 signaling. Am J Physiol Lung Cell Mol Physiol 315(2):L241–L247
Pruitt K, Der CJ (2001) Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett 171(1):1–10
Hirayama M et al (2020) FTO demethylates cyclin D1 mRNA and controls cell-cycle progression. Cell Rep 31(1):107464
Ho JY et al (2017) MicroRNA-382-5p aggravates breast cancer progression by regulating the RERG/Ras/ERK signaling axis. Oncotarget 8(14):22443–22459
Edward Zhou X, Melcher K, Eric Xu H (2019) Structural biology of G protein-coupled receptor signaling complexes. Protein Sci 28(3):487–501
Bugaj LJ et al (2018) Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science. https://doi.org/10.1126/science.aao3048
Walker JA, Upadhyaya M (2018) Emerging therapeutic targets for neurofibromatosis type 1. Expert Opin Ther Targets 22(5):419–437
Mund JA et al (2020) Genetic disruption of the small GTPase RAC1 prevents plexiform neurofibroma formation in mice with neurofibromatosis type 1. J Biol Chem 295(29):9948–9958
Dammann R et al (2005) The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol 20(2):645–663
Tong JH et al (2010) Putative tumour-suppressor gene DAB2 is frequently down regulated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer 10:253
Degirmenci U, Wang M, Hu J (2020) Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 9(1):198
Neel NF et al (2011) The RalGEF-Ral effector signaling network: the road less travelled for anti-Ras drug discovery. Genes Cancer 2(3):275–287
Guin S, Theodorescu D (2015) The RAS-RAL axis in cancer: evidence for mutation-specific selectivity in non-small cell lung cancer. Acta Pharmacol Sin 36(3):291–297
Feig LA (2003) Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol 13(8):419–425
Bodemann BO, White MA (2008) Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer 8(2):133–140
Seibold M et al (2020) RAL GTPases mediate multiple myeloma cell survival and are activated independently of oncogenic RAS. Haematologica 105(9):2316–2326
Shirakawa R, Horiuchi H (2015) Ral GTPases: crucial mediators of exocytosis and tumourigenesis. J Biochem 157(5):285–299
Masuda H et al (2012) Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat 136(2):331–345
Moghadam AR et al (2017) Ral signaling pathway in health and cancer. Cancer Med 6(12):2998–3013
Thies KA et al (2021) The small G-protein RalA promotes progression and metastasis of triple-negative breast cancer. Breast Cancer Res 23(1):65
Ghoroghi S et al (2021) Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife 10:e61539
Singh N et al (2014) Differential expression of bitter taste receptors in non-cancerous breast epithelial and breast cancer cells. Biochem Biophys Res Commun 446(2):499–503
Bum-Erdene K et al (2020) Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases. Proc Natl Acad Sci USA 117(13):7131–7139
Yan C et al (2014) Discovery and characterization of small molecules that target the GTPase Ral. Nature 515(7527):443–447
Halgren T (2007) New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 69(2):146–148
Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129(5):865–877
Maertens O, Cichowski K (2014) An expanding role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv Biol Regul 55:1–14
Guin S et al (2013) Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J Thorac Oncol 8(12):1492–1501
Saito R et al (2013) Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene 32(7):894–902
Smith SC et al (2006) The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res 66(4):1917–1922
Gilca M, Dragos D (2017) Extraoral taste receptor discovery: new light on ayurvedic pharmacology. Evid Based Complement Altern Med 2017:5435831
Slack JP et al (2010) Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr Biol 20(12):1104–1109
Singh N et al (2020) Chemosensory bitter taste receptors T2R4 and T2R14 activation attenuates proliferation and migration of breast cancer cells. Mol Cell Biochem 465(1–2):199–214
Pydi SP et al (2012) Recent advances in structure and function studies on human bitter taste receptors. Curr Protein Pept Sci 13(6):501–508
Pydi SP, Bhullar RP, Chelikani P (2012) Constitutively active mutant gives novel insights into the mechanism of bitter taste receptor activation. J Neurochem 122(3):537–544
Upadhyaya JD et al (2016) The pharmacochaperone activity of quinine on bitter taste receptors. PLoS ONE 11(5):e0156347
Delire B, Starkel P (2015) The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest 45(6):609–623
Roberts PJ, Der CJ (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26(22):3291–3310
Yap TA et al (2008) Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol 8(4):393–412
Gentry LR et al (2015) Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of Ral GTPases. J Biol Chem 290(37):22851–22861
Sidhu C et al (2017) Regulation of Rac1 GTPase activity by quinine through G-protein and bitter taste receptor T2R4. Mol Cell Biochem 426(1–2):129–136
Martin TD, Der CJ (2012) Differential involvement of RalA and RalB in colorectal cancer. Small GTPases 3(2):126–130
Clough RR, Sidhu RS, Bhullar RP (2002) Calmodulin binds RalA and RalB and is required for the thrombin-induced activation of Ral in human platelets. J Biol Chem 277(32):28972–28980
Barry SR, Bernal J (1993) Antimalarial drugs inhibit calcium-dependent backward swimming and calcium currents in Paramecium calkinsi. J Comp Physiol A 172(4):457–466
Acknowledgements
This work was supported by research funds from the Dr. Gerald Niznick College of Dentistry, University of Manitoba to R.P.B.
Author information
Authors and Affiliations
Contributions
VB designed and performed experiments, analyzed data, and contributed to figure preparation and manuscript writing; LE designed the study, analyzed data, and contributed to manuscript and figure preparation; RPB designed the study, analyzed data, and edited and finalized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest. Co-authors have read the manuscript and agreed to submission to MCB journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhatia, V., Esmati, L. & Bhullar, R.P. Regulation of Ras p21 and RalA GTPases activity by quinine in mammary epithelial cells. Mol Cell Biochem 479, 567–577 (2024). https://doi.org/10.1007/s11010-023-04725-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04725-z