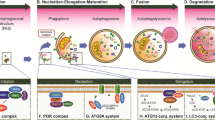
Abstract
Autophagy is the process of recycling and utilization of degraded organelles and macromolecules in the cell compartments formed during the fusion of autophagosomes with lysosomes. During autophagy induction the healthy and tumor cells adapt themselves to harsh conditions such as cellular stress or insufficient supply of nutrients in the cell environment to maintain their homeostasis. Autophagy is currently seen as a form of programmed cell death along with apoptosis and necroptosis. In recent years multiple studies have considered the autophagy as a potential mechanism of anticancer therapy in malignant glioma. Although, subsequent steps in autophagy development are known and well-described, on molecular level the mechanism of autophagosome initiation and maturation using autophagy-related proteins is under investigation. This article reviews current state about the mechanism of autophagy, its molecular pathways and the most recent studies on roles of autophagy-related proteins and their isoforms in glioma progression and its treatment.


Similar content being viewed by others
References
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326. https://doi.org/10.1016/j.cell.2010.01.028
Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G (2017) Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov 16:487–511. https://doi.org/10.1038/nrd.2017.22
Mijaljica D, Prescott M, Devenish RJ (2011) Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7:673–682. https://doi.org/10.4161/auto.7.7.14733
Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221:3–12. https://doi.org/10.1002/path.2697
Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. https://doi.org/10.1146/annurev-cellbio-092910-154005
Schneider JL, Cuervo AM (2014) Autophagy and human disease: emerging themes. Curr Opin Genet Dev 26:16–23
Chu CT (2008) Eaten alive: autophagy and neuronal cell death after hypoxia-ischemia. Am J Pathol 172:284–287. https://doi.org/10.2353/ajpath.2008.071064
Cuervo AM (2004) Autophagy: in sickness and in health. Trends Cell Biol 14:70–77. https://doi.org/10.1016/j.tcb.2003.12.002
Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2010) Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy 6:301–303. https://doi.org/10.4161/auto.6.2.11134
Zachari M, Ganley IG (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61:585–596. https://doi.org/10.1042/EBC20170021
Meng D, Frank AR, Jewell JL (2018) mTOR signaling in stem and progenitor cells. Development. https://doi.org/10.1242/dev.152595
Carroll B, Maetzel D, Maddocks OD, Otten G, Ratcliff M, Smith GR, Dunlop EA, Passos JF, Davies OR, Jaenisch R, Tee AR, Sarkar S, Korolchuk VI (2016) Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. Elife. https://doi.org/10.7554/eLife.11058
Cao Y, Klionsky DJ (2007) Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 17:839–849. https://doi.org/10.1038/cr.2007.78
Sun Q, Fan W, Zhong Q (2009) Regulation of Beclin 1 in autophagy. Autophagy 5:713–716. https://doi.org/10.4161/auto.5.5.8524
Carew JS, Kelly KR, Nawrocki ST (2012) Autophagy as a target for cancer therapy: new developments. Cancer Manag Res 4:357–365. https://doi.org/10.2147/CMAR.S26133
Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B (1999) Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59:59–65. https://doi.org/10.1006/geno.1999.5851
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676. https://doi.org/10.1038/45257
Avalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF (2014) Tumor suppression and promotion by autophagy. Biomed Res Int 2014:603980. https://doi.org/10.1155/2014/603980
Pracharova J, Radosova Muchova T, Dvorak Tomastikova E, Intini FP, Pacifico C, Natile G, Kasparkova J, Brabec V (2016) Anticancer potential of a photoactivated transplatin derivative containing the methylazaindole ligand mediated by ROS generation and DNA cleavage. Dalton Trans 45:13179–13186. https://doi.org/10.1039/c6dt01467d
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42. https://doi.org/10.1016/j.cell.2007.12.018
Yang X, Yu DD, Yan F, Jing YY, Han ZP, Sun K, Liang L, Hou J, Wei LX (2015) The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci 5:14. https://doi.org/10.1186/s13578-015-0005-2
Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC (2011) Pancreatic cancers require autophagy for tumor growth. Genes Dev 25:717–729. https://doi.org/10.1101/gad.2016111
Zhu L, Fu X, Yuan C, Jiang X, Zhang G (2018) Induction of oxidative DNA damage in bovine herpesvirus 1 infected bovine kidney cells (MDBK cells) and human tumor cells (A549 cells and U2OS cells). Viruses. https://doi.org/10.3390/v10080393
Auten RL, Davis JM (2009) Oxygen toxicity and reactive oxygen species: the devil is in the details. Pediatr Res 66:121–127. https://doi.org/10.1203/PDR.0b013e3181a9eafb
Dewhirst MW, Cao Y, Moeller B (2008) Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer 8:425–437. https://doi.org/10.1038/nrc2397
Liu T, Wu L, Wang D, Wang H, Chen J, Yang C, Bao J, Wu C (2016) Role of reactive oxygen species-mediated MAPK and NF-kappaB activation in polygonatum cyrtonema lectin-induced apoptosis and autophagy in human lung adenocarcinoma A549 cells. J Biochem 160:315–324. https://doi.org/10.1093/jb/mvw040
Hui KF, Yeung PL, Chiang AK (2016) Induction of MAPK- and ROS-dependent autophagy and apoptosis in gastric carcinoma by combination of romidepsin and bortezomib. Oncotarget 7:4454–4467. https://doi.org/10.18632/oncotarget.6601
Zhang SY, Li XB, Hou SG, Sun Y, Shi YR, Lin SS (2016) Cedrol induces autophagy and apoptotic cell death in A549 non-small cell lung carcinoma cells through the P13K/Akt signaling pathway, the loss of mitochondrial transmembrane potential and the generation of ROS. Int J Mol Med 38:291–299. https://doi.org/10.3892/ijmm.2016.2585
Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 28:1341–1350. https://doi.org/10.1038/emboj.2009.80
Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, Kusewitt D, Mills GB, Kastan MB, Walker CL (2010) ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA 107:4153–4158. https://doi.org/10.1073/pnas.0913860107
Ryter SW, Mizumura K, Choi AM (2014) The impact of autophagy on cell death modalities. Int J Cell Biol 2014:502676. https://doi.org/10.1155/2014/502676
Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM, Narita M (2009) Autophagy mediates the mitotic senescence transition. Genes Dev 23:798–803. https://doi.org/10.1101/gad.519709
Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40:280–293. https://doi.org/10.1016/j.molcel.2010.09.023
Frisch SM, Francis H (1994) Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol 124:619–626. https://doi.org/10.1083/jcb.124.4.619
Lisanti MP, Martinez-Outschoorn UE, Chiavarina B, Pavlides S, Whitaker-Menezes D, Tsirigos A, Witkiewicz A, Lin Z, Balliet R, Howell A, Sotgia F (2010) Understanding the “lethal” drivers of tumor-stroma co-evolution: emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Biol Ther 10:537–542. https://doi.org/10.4161/cbt.10.6.13370
Silva LM, Jung JU (2013) Modulation of the autophagy pathway by human tumor viruses. Semin Cancer Biol 23:323–328. https://doi.org/10.1016/j.semcancer.2013.05.005
Avia M, Rojas JM, Miorin L, Pascual E, Van Rijn PA, Martin V, Garcia-Sastre A, Sevilla N (2019) Virus-induced autophagic degradation of STAT2 as a mechanism for interferon signaling blockade. EMBO Rep 20:e48766. https://doi.org/10.15252/embr.201948766
Li Y, Hu B, Ji G, Zhang Y, Xu C, Lei J, Ding C, Zhou J (2020) Cytoplasmic cargo receptor p62 inhibits avibirnavirus replication by mediating autophagic degradation of viral protein VP2. J Virol. https://doi.org/10.1128/JVI.01255-20
Muscolino E, Schmitz R, Loroch S, Caragliano E, Schneider C, Rizzato M, Kim YH, Krause E, Juranic Lisnic V, Sickmann A, Reimer R, Ostermann E, Brune W (2020) Herpesviruses induce aggregation and selective autophagy of host signalling proteins NEMO and RIPK1 as an immune-evasion mechanism. Nat Microbiol 5:331–342. https://doi.org/10.1038/s41564-019-0624-1
zur Hausen H (1991) Viruses in human cancers. Science 254:1167–1173. https://doi.org/10.1126/science.1659743
Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, Villano JL (2014) Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prevent 23:1985–1996. https://doi.org/10.1158/1055-9965.EPI-14-0275
D’Alessio A, Proietti G, Sica G, Scicchitano BM (2019) Pathological and molecular features of glioblastoma and its peritumoral tissue. Cancers (Basel). https://doi.org/10.3390/cancers11040469
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO, European Organisation for R, Treatment of Cancer Brain T, Radiation Oncology G and National Cancer Institute of Canada Clinical Trials G (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466. https://doi.org/10.1016/S1470-2045(09)70025-7
Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee ShU (2017) Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prevent 18:3–9. https://doi.org/10.22034/APJCP.2017.18.1.3
Kaza N, Kohli L, Roth KA (2012) Autophagy in brain tumors: a new target for therapeutic intervention. Brain Pathol 22:89–98. https://doi.org/10.1111/j.1750-3639.2011.00544.x
Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E (2001) The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 276:35243–35246. https://doi.org/10.1074/jbc.C100319200
Chen J, Zeng F, Forrester SJ, Eguchi S, Zhang MZ, Harris RC (2016) Expression and function of the epidermal growth factor receptor in physiology and disease. Physiol Rev 96:1025–1069. https://doi.org/10.1152/physrev.00030.2015
Vescovo T, Pagni B, Piacentini M, Fimia GM, Antonioli M (2020) Regulation of autophagy in cells infected with oncogenic human viruses and its impact on cancer development. Front Cell Dev Biol 8:47. https://doi.org/10.3389/fcell.2020.00047
Kudchodkar SB, Levine B (2009) Viruses and autophagy. Rev Med Virol 19:359–378. https://doi.org/10.1002/rmv.630
Henderson V, Smith B, Burton LJ, Randle D, Morris M, Odero-Marah VA (2015) Snail promotes cell migration through PI3K/AKT-dependent Rac1 activation as well as PI3K/AKT-independent pathways during prostate cancer progression. Cell Adh Migr 9:255–264. https://doi.org/10.1080/19336918.2015.1013383
Mahajan-Thakur S, Bien-Moller S, Marx S, Schroeder H, Rauch BH (2017) Sphingosine 1-phosphate (S1P) signaling in glioblastoma multiforme-A systematic review. Int J Mol Sci. https://doi.org/10.3390/ijms18112448
Gonzalez E, McGraw TE (2009) The Akt kinases: isoform specificity in metabolism and cancer. Cell Cycle 8:2502–2508. https://doi.org/10.4161/cc.8.16.9335
Endersby R, Zhu X, Hay N, Ellison DW, Baker SJ (2011) Nonredundant functions for Akt isoforms in astrocyte growth and gliomagenesis in an orthotopic transplantation model. Cancer Res 71:4106–4116. https://doi.org/10.1158/0008-5472.CAN-10-3597
Wang Q, Chen X, Hay N (2017) Akt as a target for cancer therapy: more is not always better (lessons from studies in mice). Br J Cancer 117:159–163. https://doi.org/10.1038/bjc.2017.153
Joy A, Kapoor M, Georges J, Butler L, Chang Y, Li C, Crouch A, Smirnov I, Nakada M, Hepler J, Marty M, Feuerstein BG (2016) The role of AKT isoforms in glioblastoma: AKT3 delays tumor progression. J Neurooncol 130:43–52. https://doi.org/10.1007/s11060-016-2220-z
Kaley TJ, Panageas KS, Mellinghoff IK, Nolan C, Gavrilovic IT, DeAngelis LM, Abrey LE, Holland EC, Lassman AB (2019) Phase II trial of an AKT inhibitor (perifosine) for recurrent glioblastoma. J Neurooncol 144:403–407. https://doi.org/10.1007/s11060-019-03243-7
Ronellenfitsch MW, Zeiner PS, Mittelbronn M, Urban H, Pietsch T, Reuter D, Senft C, Steinbach JP, Westphal M, Harter PN (2018) Akt and mTORC1 signaling as predictive biomarkers for the EGFR antibody nimotuzumab in glioblastoma. Acta Neuropathol Commun 6:81. https://doi.org/10.1186/s40478-018-0583-4
Kaley TJ, Panageas KS, Pentsova EI, Mellinghoff IK, Nolan C, Gavrilovic I, DeAngelis LM, Abrey LE, Holland EC, Omuro A, Lacouture ME, Ludwig E, Lassman AB (2020) Phase I clinical trial of temsirolimus and perifosine for recurrent glioblastoma. Ann Clin Transl Neurol 7:429–436. https://doi.org/10.1002/acn3.51009
Bozic M, van den Bekerom L, Milne BA, Goodman N, Roberston L, Prescott AR, Macartney TJ, Dawe N, McEwan DG (2020) A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep 21:e48412. https://doi.org/10.15252/embr.201948412
Fu Y, Huang Z, Hong L, Lu JH, Feng D, Yin XM, Li M (2019) Targeting ATG4 in cancer therapy. Cancers (Basel). https://doi.org/10.3390/cancers11050649
Agrotis A, Pengo N, Burden JJ, Ketteler R (2019) Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 15:976–997. https://doi.org/10.1080/15548627.2019.1569925
Bortnik S, Choutka C, Horlings HM, Leung S, Baker JH, Lebovitz C, Dragowska WH, Go NE, Bally MB, Minchinton AI, Gelmon KA, Gorski SM (2016) Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 7:66970–66988. https://doi.org/10.18632/oncotarget.11408
Tran E, Chow A, Goda T, Wong A, Blakely K, Rocha M, Taeb S, Hoang VC, Liu SK, Emmenegger U (2013) Context-dependent role of ATG4B as target for autophagy inhibition in prostate cancer therapy. Biochem Biophys Res Commun 441:726–731. https://doi.org/10.1016/j.bbrc.2013.10.117
Mao JJ, Wu LX, Wang W, Ye YY, Yang J, Chen H, Yang QF, Zhang XY, Wang B, Chen WX (2018) Nucleotide variation in ATG4A and susceptibility to cervical cancer in Southwestern Chinese women. Oncol Lett 15:2992–3000. https://doi.org/10.3892/ol.2017.7663
Wen ZP, Zeng WJ, Chen YH, Li H, Wang JY, Cheng Q, Yu J, Zhou HH, Liu ZZ, Xiao J, Chen XP (2019) Knockdown ATG4C inhibits gliomas progression and promotes temozolomide chemosensitivity by suppressing autophagic flux. J Exp Clin Cancer Res 38:298. https://doi.org/10.1186/s13046-019-1287-8
Gil J, Ramsey D, Pawlowski P, Szmida E, Leszczynski P, Bebenek M, Sasiadek MM (2018) The influence of tumor microenvironment on ATG4D gene expression in colorectal cancer patients. Med Oncol 35:159. https://doi.org/10.1007/s12032-018-1220-6
Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW (2005) Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 280:18283–18290. https://doi.org/10.1074/jbc.M413957200
Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 148:465–480. https://doi.org/10.1083/jcb.148.3.465
Abdul Rahim SA, Dirkse A, Oudin A, Schuster A, Bohler J, Barthelemy V, Muller A, Vallar L, Janji B, Golebiewska A, Niclou SP (2017) Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer 117:813–825. https://doi.org/10.1038/bjc.2017.263
Costa JR, Prak K, Aldous S, Gewinner CA, Ketteler R (2016) Autophagy gene expression profiling identifies a defective microtubule-associated protein light chain 3A mutant in cancer. Oncotarget 7:41203–41216. https://doi.org/10.18632/oncotarget.9754
Wang N, Tan HY, Li S, Feng Y (2017) Atg9b deficiency suppresses autophagy and potentiates endoplasmic reticulum stress-associated hepatocyte apoptosis in hepatocarcinogenesis. Theranostics 7:2325–2338. https://doi.org/10.7150/thno.18225
Moazeni-Roodi A, Tabasi F, Ghavami S, Hashemi M (2019) Investigation of ATG16L1 rs2241880 polymorphism with cancer risk: a meta-analysis. Medicina (Kaunas). https://doi.org/10.3390/medicina55080425
Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, Gentle D, Catchpoole D, Maher ER, Pfeifer GP, Latif F (2010) A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer 9:44. https://doi.org/10.1186/1476-4598-9-44
Frolov A, Evans IM, Li N, Sidlauskas K, Paliashvili K, Lockwood N, Barrett A, Brandner S, Zachary IC, Frankel P (2016) Imatinib and Nilotinib increase glioblastoma cell invasion via Abl-independent stimulation of p130Cas and FAK signalling. Sci Rep 6:27378. https://doi.org/10.1038/srep27378
Koukourakis MI, Kalamida D, Giatromanolaki A, Zois CE, Sivridis E, Pouliliou S, Mitrakas A, Gatter KC, Harris AL (2015) Autophagosome proteins LC3A, LC3B and LC3C have distinct subcellular distribution kinetics and expression in cancer cell lines. PLoS ONE 10:e0137675. https://doi.org/10.1371/journal.pone.0137675
Aoki H, Kondo Y, Aldape K, Yamamoto A, Iwado E, Yokoyama T, Hollingsworth EF, Kobayashi R, Hess K, Shinojima N, Shingu T, Tamada Y, Zhang L, Conrad C, Bogler O, Mills G, Sawaya R, Kondo S (2008) Monitoring autophagy in glioblastoma with antibody against isoform B of human microtubule-associated protein 1 light chain 3. Autophagy 4:467–475. https://doi.org/10.4161/auto.5668
Giatromanolaki A, Sivridis E, Mitrakas A, Kalamida D, Zois CE, Haider S, Piperidou C, Pappa A, Gatter KC, Harris AL, Koukourakis MI (2014) Autophagy and lysosomal related protein expression patterns in human glioblastoma. Cancer Biol Ther 15:1468–1478. https://doi.org/10.4161/15384047.2014.955719
Zhang J (2015) Teaching the basics of autophagy and mitophagy to redox biologists–mechanisms and experimental approaches. Redox Biol 4:242–259. https://doi.org/10.1016/j.redox.2015.01.003
Pirtoli L, Cevenini G, Tini P, Vannini M, Oliveri G, Marsili S, Mourmouras V, Rubino G, Miracco C (2009) The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy 5:930–936. https://doi.org/10.4161/auto.5.7.9227
Kaverina NV, Kadagidze ZG, Borovjagin AV, Karseladze AI, Kim CK, Lesniak MS, Miska J, Zhang P, Baryshnikova MA, Xiao T, Ornelles D, Cobbs C, Khramtsov A, Ulasov IV (2018) Tamoxifen overrides autophagy inhibition in Beclin-1-deficient glioma cells and their resistance to adenovirus-mediated oncolysis via upregulation of PUMA and BAX. Oncogene. https://doi.org/10.1038/s41388-018-0395-9
Padmakrishnan CJ, Easwer HV, Vijayakurup V, Menon GR, Nair S, Gopala S (2019) High LC3/beclin expression correlates with poor survival in glioma: a definitive role for autophagy as evidenced by in vitro autophagic flux. Pathol Oncol Res 25:137–148. https://doi.org/10.1007/s12253-017-0310-7
Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, Pirtoli L, Miracco C (2013) microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther 14:574–586. https://doi.org/10.4161/cbt.24597
Graf MR, Jia W, Loria RM (2007) The neuro-steroid, 3beta androstene 17alpha diol exhibits potent cytotoxic effects on human malignant glioma and lymphoma cells through different programmed cell death pathways. Br J Cancer 97:619–627. https://doi.org/10.1038/sj.bjc.6603894
Huynh PN, Loria RM (1997) Contrasting effects of alpha- and beta-androstenediol on oncogenic myeloid cell lines in vitro. J Leukoc Biol 62:258–267. https://doi.org/10.1002/jlb.62.2.258
Huynh PN, Carter WH Jr, Loria RM (2000) 17 alpha androstenediol inhibition of breast tumor cell proliferation in estrogen receptor-positive and -negative cell lines. Cancer Detect Prev 24:435–444
Acknowledgements
This work was financed by the Ministry of Science and Higher Education of the Russian Federation within the framework of state support for the creation and development of World-Class Research Centers "Digital biodesign and personalized healthcare" No. 075-15-2020-926.
Funding
This work is supported by Russian Science Foundation under grant No. 21-15-00213 (IU, description of molecular mechanisms of autophagy).
Author information
Authors and Affiliations
Contributions
EB: Conception, design, writing – review & editing. RKK: revising the manuscript, drafting, editing. IVU: supervise, critically edited and approved find draft for submission. PT: Revising and approve the final manuscript for submission. IK-editing manuscript; RM: revising and approve the manuscript to be published. AA: Revising critically and approve the final manuscript for submission.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animal, performed by any of the author.
Data availability
All data generated or analysed during this study are included in this published article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Belyaeva, E., Kharwar, R.K., Ulasov, I.V. et al. Isoforms of autophagy-related proteins: role in glioma progression and therapy resistance. Mol Cell Biochem 477, 593–604 (2022). https://doi.org/10.1007/s11010-021-04308-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04308-w