Abstract
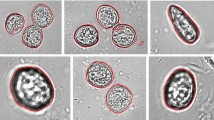
Microscopic phytoplankton segmentation is an important part of water quality assessment. The segmentation of microscopic phytoplankton still faces challenges for computer vision, such as being affected by background impurities and requiring a large number of manual annotation. In this paper, the characteristics of phytoplankton emitting fluorescence under excitation light were utilized to segment and annotate phytoplankton contours by fusing fluorescence images and bright field images. Morphological operations were used to process microscopic fluorescence images to obtain the initial contours of phytoplankton. Then, microscopic bright field images were processed by Active Contour to fine tune the contours. Seven algae species were selected as the experimental objects. Compared with manually labeling the contour in LabelMe, the recall, precision, FI score and IOU of the proposed segmentation method are 85.3%, 84.5%, 84.7%, and 74.6%, respectively. Mask-RCNN was used to verify the correctness of labels annotated by the proposed method. The average recall, precision, F1 score and IOU are 97.0%, 86.5%, 91.1%, and 84.2%, respectively, when the Mask-RCNN is trained with the proposed automatic labeling method. And the results corresponding to manual labeling are 95.3%, 86.1%, 90.3%, and 82.8% respectively. The experimental results show that the proposed method can segment the phytoplankton microscopic image accurately, and the automatically annotated contour data has the same effect as the manually annotated contour data in Mask-RCNN, which greatly reduces the manual annotation workload.








Similar content being viewed by others
Data Availability
The data that support outcomes of this study are available on reasonable request from the corresponding author.
References
Suthers I, Rissik D (2009) Plankton: A guide to their ecology and monitoring for water quality (CSIRO Publishing)
Coltelli P, Barsanti L, Evangelista V, Frassantio AM, Gualtieri P (2014) Water monitoring: automated and real time identification and classification of algae using digital microscopy. Environ Sci Processes Impacts 16:2656
Barsanti L, Evangelista V, Gualtieri P (2021) Water monitoring by means of digital microscopy identification and classification of microalgae. Environ Sci Processes Impacts 23:1443
Jalba AC, Wilkinson MH, Roerdink JBTM (2004) Automatic segmentation of diatom images for classification. Microsc Res Tech 65:72
Bi HS, Guo ZH, Benfield MC, Fan CL, Ford M, Shahrestani S, Sieracki JM (2015) A semi-automated image analysis procedure for in situ plankton imaging systems. PLoS ONE 10:e0127121
Natchimuthu S, Chinnaraj P, Parthasarathy S, Senthil K (2013) Automatic identification of algal community from microscopic images. Bioinf Biol Insights 7:327
Dimitrovski I, Kocev D, Loskovska S, Dzeroski S (2012) Hierachical classification of diatom images using ensembles of predictive clustering trees. Eco Inform 7:19
Zheng HY, Wang N, Yu ZB, Gu ZR, Zheng B (2017) Robust and automatic cell detection and segmentation from microscopic images of non-setae phytoplankton species. IET Image Proc 11:1077
Valentini N, Balouin Y (2020) Assessment of a smartphone-based camera system for coastal image segmentation and sargassum monitoring. J Mar Sci Eng 8:23
Li Y, Di J, Ren L, Zhao J (2021) Deep-learning-based prediction of living cells mitosis via quantitative phase microscopy. Chin Opt Lett 19:051701
Liang Z, Wang J, Xiao G, Zeng L (2022) FAANET: feature-aligned attention networks for real-time multiple object tracking in UAV videos. Chin Opt Lett 20:081101
He K, Gkioxari G, Dollár P, Girshick R (2020) Mask R-CNN. IEEE Trans Pattern Anal Mach Intell 42:386
Ruiz-Santaquiteria J, Bueno G, Deniz O, Vallez N, Cristobal G (2020) Semantic versus instance segmentation in microscopic algae detection. Eng Appl Artif Intell 87:103271
Suh S, Park Y, Ko K, Yang S, Ahn J, Shin JK, Kim S (2021) Weighted Mask R-CNN for Improving Adjacent Boundary Segmentation. J Sens 2021:8872947
Baek SS, Pyo J, Pachepsky Y, Park Y, Ligaray M, Ahn CY, Kim YH, Chun JA, Cho KH (2020) Identification and enumeration of cyanobacteria species using a deep neural network. Ecol Indic 115:106395
Russell BC, Torralba A, Murphy KP, Freeman WT (2008) LabelMe: A Database and Web-Based Tool for Image Annotation. Int J Comput Vision 77:157
Dutta A, Zissermann A (2019) The VIA Annotation Software for Images, Audio and Video, in the 27th ACM international conference on multimedia, p 2276
Mäkinen S, Skogstrӧm H, Laaksonen E, Mikkonen T (2021) Who needs MLOps: What data scientists seek to accomplish and how can MLOps help? In: IEEE/ACM 1st Workshop on AI Engineering - Software Engineering for AI, p 109
Gelzinis A, Verikas A, Vaiciukynas E, Bacauskiene M (2015) A novel technique to extract accurate cell contours applied for segmentation of phytoplankton images. Mach Vis Appl 26:305–315
Zhang ZY, Yin XL, Yan ZY (2021) Rapid data annotation for sand-like granular instance segmentation using mask-RCNN. Autom Constr 133:103994
Yin GF, Zhao NJ, Hu L, Yu XY, Shi CY, Xiao X, Fang F, Duan JB, Gan TT, Zhang YJ, Liu JG, Liu WQ (2014) Classified measurement of phytoplankton based on characteristic fluorescence of photosynthetic pigments. Acta Opt Sin 34:0930005
Jia RQ, Yin GF, Zhao NJ, Xu M, Huang P, Liang TH, He QF, Chen XW, Gan TT, Zhang XL, Ma MJ (2022) Registration Method of Microscopic Bright Field and Fluorescence Synchronous Measurement Images of Phytoplankton Cells. Chin J Lasers 49:2407202
Gonzalez RC, Woods RE (2008) Digital Image Processing, 3rd ed.” (Prentice Hall)
Funding
This work was supported by the Science and Technology Major Special Project of Anhui Province (202203a07020002,202003a07020007); the National Key Research and Development Program of China (2021YFC3200100); the National Natural Science Foundation of China (61875207, 62005001).
Author information
Authors and Affiliations
Contributions
Writing, renqing Jia; Funding acquisition, Gaofang Yin, Xiaoling Zhang, and Nanjing Zhao; Supervision, Qianfeng He, and Nanjing Zhao; Resources, Min Xu, Xiang Hu, Peng Huang, and Tianhong Liang; English polishing, Xiaowei Chen. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ehical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All the authors agree to publish this article.
Competing Interests
The authors declare no conflict of interest.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jia, R., Yin, G., Zhao, N. et al. Phytoplankton Image Segmentation and Annotation Method Based on Microscopic Fluorescence. J Fluoresc (2023). https://doi.org/10.1007/s10895-023-03515-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10895-023-03515-6