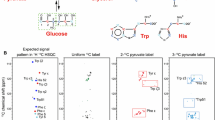
Abstract
Cell-free protein synthesis using eCells allows production of amino acids from inexpensive 13C-labelled precursors. We show that the metabolic pathway converting pyruvate, glucose and erythrose into aromatic amino acids is maintained in eCells. Judicious choice of 13C-labelled starting material leads to proteins, where the sidechains of aromatic amino acids display [13C,1H]-HSQC cross-peaks free of one-bond 13C–13C couplings. Selective 13C-labelling of tyrosine and phenylalanine residues is achieved simply by using different compositions of the reaction buffers.
Similar content being viewed by others
Introduction
The NMR spectroscopic analysis of proteins of increasing molecular weight is hindered by cross-peak overlap. Selective isotope-labelling presents an elegant way for improving the spectral resolution (Verardi et al. 2012) with site-selective 13C and 2H labelling delivering greatly simplified protein NMR spectra, which benefit the analysis of large proteins (Takeuchi et al. 2007; Kainosho and Güntert 2009; Takeda et al. 2010). As a drawback, the cost of sample preparation with isotope-labelled amino acids can be high (Gell et al. 2017). The present work presents a new strategy for selective 13C-labelling of aromatic amino acids that uses inexpensive 13C-labelled precursors to direct 13C labels into specific positions of the aromatic side chains of phenylalanine (Phe), tryptophan (Trp) and tyrosine (Tyr) in a cell-free protein synthesis (CFPS) system. NMR spectroscopy offers powerful means to assess the dynamics of aromatic amino acid side chains, and site-selective 13C-labels present particularly convenient probes (Akke and Weininger 2023).
Due to large one-bond 13C–13C couplings (1JCC) and limited chemical shift dispersion between the δ and ε CH groups of Tyr and δ, ε and ζ CH groups of Phe (Williams et al. 2015), the [13C,1H]-HSQC spectra of the aromatic rings of Phe, Trp and Tyr residues are prone to spectral overlap, when the proteins are produced with uniform 13C-labelling (Torizawa et al. 2005; Milbradt et al. 2015). Furthermore, 1JCC couplings of aromatic side chains are large and hinder relaxation measurements to probe for conformational changes (Teilum et al. 2006; Kasinath et al. 2013). 1JCC couplings also limit the gain in sensitivity and resolution that can be obtained from aromatic TROSY experiments (Pervushin et al. 1998; Milbradt et al. 2015). This situation has triggered the development of various atom-specific labelling methods to reduce the complexity of the 13C-NMR spectra of aromatic amino acids by eliminating 1JCC couplings (Schörghuber et al. 2018). As the chemical synthesis of suitably 13C-labelled amino acids is expensive, a more cost-efficient approach is to harness the anabolic pathways of in vivo amino acid synthesis to produce specifically labelled protein from relatively inexpensive labelled precursors. In the case of Phe and Tyr, E. coli cell cultures in minimal media supplied with 13C-labelled α-ketoacids as late-stage precursors were shown to produce selectively labelled protein in vivo without cross-labelling (Lichtenecker et al. 2013; Lichtenecker 2014). Selectively 13C-labelled pyruvate and erythrose, which are earlier precursors of aromatic amino acids, have also been used successfully to label single δ-carbon sites in Phe and Tyr and the ε3-carbon in Trp (Kasinath et al. 2013).
In vivo labelling strategies are prone to isotope scrambling, which can be suppressed by limiting the time of bacterial cell growth after induction to prevent precursor recycling (Kurauskas et al. 2017). In an extension of this approach, selected amino acids can be “unlabelled” by their provision in the growth medium in their standard form, i.e., at natural isotopic abundance (Rasia et al. 2012; Lacabanne et al. 2018).
To obtain the protein quantities required for NMR spectroscopy, in vivo protein expression requires significant amounts of the isotope-labelled amino acids or precursors, whereas contemporary cell-free protein synthesis systems incorporate amino acids in much higher yield (Torizawa et al. 2004). Unfortunately, many of the bacterial enzymes are rendered nonfunctional during the preparation of cell extracts, so that the less expensive isotope labelling approaches that depend on multi-step anabolic pathways are inaccessible to conventional CFPS (Linser et al. 2014). As replenishing the cell-free extract with the requisite enzymes would be costly and unpractical, we explored the use of the recently developed eCell system for protein production under CFPS conditions. eCells are bacterial cells coated with layers of differently charged polyionic polymers and cell walls made porous by the expression of endolysin (Van Raad and Huber 2021). The pores of eCells leak proteins only very slowly over a period of hours, whereas low-molecular weight compounds readily exchange between the intra- and extracellular space. eCells can therefore be compared with traditional CFPS systems that use dialysis membranes separating inner and outer buffer solutions (Apponyi et al. 2008). Importantly, the mild preparation conditions of eCells maintain the activity of bacterial anabolic enzymes much better than cell extracts. In previous work we observed that, besides maintaining the entire repertoire of transcription/translation components, eCells also preserve the enzymes required for the biosynthesis of valine and leucine from pyruvate and glucose (Van Raad et al. 2023). The present work shows that eCells likewise preserve the fully functional complement of biosynthetic E. coli enzymes required for the synthesis of Phe, Trp and Tyr and therefore allow the production of proteins with site-specifically 13C-labelled aromatic amino acids from inexpensive precursors.
The bacterial biosynthetic pathways of aromatic amino acid synthesis (Fig. 1) show that pyruvate and erythrose are precursors that lead to the final amino acid product via a limited number of intermediates. The combination of phosphoenol pyruvate with erythrose phosphate produces Phe, Trp and Tyr, indicating that selective 13C-labelling of the δ and ε positions of Tyr/Phe and the δ2 position of Trp can be achieved in eCells by the use of 13C-labelled pyruvate and erythrose, in analogy to the situation in vivo (Lundström et al. 2007). The present work shows that this is indeed the case.
Biosynthetic pathway of the aromatic amino acids tyrosine, phenylalanine and tryptophan starting from phosphoenol-pyruvate and erythrose-4-phosphate. The present work used 4-13C erythrose and 3-13C pyruvate in eCells to produce proteins with specifically 13C-labelled aromatic amino acids. Red balls mark the 13C-enriched positions and trace their fate through the pathways
Materials and methods
Plasmids
The expression vectors used for E. coli peptidyl–prolyl cis–trans isomerase B (PpiB) and ubiquitin were described previously (Van Raad and Huber 2021). They afford expression of the target proteins under control of a T7 promoter and lac operator and contain a spectinomycin resistance gene.
eCells
The production of eCells followed a previously published protocol, using E. coli autolysis Xjb(DE3)* cells, which have an endolysin gene in the genome under a PBAD promoter (Van Raad et al. 2021). The culture was induced for endolysin production at the time of inoculation with a final concentration of 3 mM arabinose. The cells were grown to OD600 0.6 and washed three times with PBS-E (phosphate-buffered saline containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 and 1 mM EDTA) pH 7.4 and resuspended in 0.25 mg/mL of chitosan in PBS-E solution. After vigorous shaking for 20 min, the cell pellet was washed with PBS-E pH 6.0 three times to remove excess chitosan and then resuspended in 0.25 mg/mL of alginate PBS-E solution and subjected to vigorous shaking for 20 min. The cells were then washed 3 times with PBS-E pH 6.0, resuspended in PBS-E pH 7.4 and directly stored at −80 oC.
CFPS
The protocol for eCell CFPS based on glucose and pyruvate has been described previously (Van Raad et al. 2023). The protocol was adapted from the phosphate recycling system published by Jewett and Swartz (2004). The CFPS buffer contained 0.9 mM UTP and CTP, 50 mM HEPES, 1.5 mM GTP, 1.5 mM ATP, 0.68 µM folinic acid, 0.64 mM cAMP, 1.7 mM DTT, 60 mM KGlu, 8 mM MgGlu, 2% v/v PEG-8000, 5 mM CoA, 33 mM pyruvate, 4 mM sodium oxalate, 0.25 mM CoA and 0.33 mM NAD. 3.5 mM of each amino acid was added except those targeted for isotope enrichment by the eCells. Na2HPO4 was omitted to promote the use of glucose for energy generation, and 4 mM sodium oxalate was used to promote the generation of ATP by the PANOx system of Jewett and Swartz (2004).
Prior to each CFPS reaction, the buffer was adjusted to pH 7.5. Frozen aliquots of encapsulated cells were thawed and the pellet immersed in CFPS buffer. Except where stated otherwise, 1.5 g eCells produced from 1 L cell culture at OD600 0.6 were suspended in 25 mL CFPS buffer. CFPS was conducted for 12 h at 37 oC in a shaker at 180 rpm. Proteins were extracted from the eCells by sonication for 2 min at 60 kHz and spinning down the resulting mixture for 15 min at 24,000 g prior to purification using His Gravitrap columns (GE Healthcare).
Labelling with 3-13C pyruvate, 2-13C glucose or with 1-13C glucose and 4-13C erythrose combined
To produce ubiquitin samples from 3-13C pyruvate for 13C-labelling of the δ positions of tyrosine and phenylalanine, dry 3-13C pyruvate was added to 300 mg eCells in 10 mL CFPS buffer at 33 mM final concentration, and Tyr or Phe were omitted from the CFPS buffer.
To test the performance of 2-13C-glucose as carbon source in the aromatic amino acid synthesis, dry 2-13C glucose was added at 30 mM final concentration to 1.5 g eCells suspended in 25 mL CFPS buffer. Tyrosine, phenylalanine and tryptophan were omitted from the amino acid mixture to allow for labelling of the ε positions of tyrosine and phenylalanine and the tryptophan δ2 position.
For increased levels of 13C labelling of PpiB, 3-13C pyruvate was added to CFPS buffers at 33 mM final concentration combined with 4-13C erythrose at 30 mM concentration. Tyrosine and phenylalanine were omitted from the CFPS reaction mixture.
For increased 13C labelling of PpiB using pyruvate and an isotopologue of glucose, 3-13C pyruvate was added to CFPS buffer at 16.5 mM final concentration combined with 1-13C glucose at 16 mM concentration. Tyrosine and phenylalanine were omitted from the reaction mixture. For all samples, 1 mM IPTG was added to the CFPS reaction mixture.
NMR spectroscopy and isotope labelling yields
[13C,1H]-HSQC spectra were recorded at 25 oC using Bruker 600 or 800 MHz NMR spectrometers equipped with TCI cryoprobes. To determine the degree of isotope enrichment, the [13C,1H]-HSQC cross-peak integrals of the labelled residues were compared with those of an internal standard of 0.1 mM 3-13C pyruvate in spectra recorded with a recovery delay of 30 s between scans.
Results
The biosynthesis of aromatic amino acids under CFPS conditions was explored using eCells made from E. coli XjB(DE3)* cells. The eCells contained the expression plasmids for ubiquitin or the E. coli prolyl cis–trans peptidyl isomerase B (PpiB). eCell CFPS supplied with 3-13C pyruvate is expected to label a single δ carbon of phenylalanine and tyrosine (Fig. 1), while for tryptophan the 13C label is expected to appear in the position of a quaternary carbon which does not produce a [13C,1H]-HSQC cross-peak.
The NMR spectra of ubiquitin confirmed these expectations. Ubiquitin contains two Phe, one Tyr and no Trp residue. As expected, two [13C,1H]-cross-peaks are observed for the sample prepared with Phe omitted and unlabelled Tyr added in the CFPS buffer (Fig. 2A), whereas a single cross-peak is observed for the sample prepared with Tyr omitted and Phe added (Fig. 2B). As anticipated, these cross-peaks show no evidence of 1JCC splittings, which are prominent in uniformly 13C-labelled samples (Fig. 2C). 300 mg eCells suspended in 10 mL buffer containing 33 mM 3-13C pyruvate were sufficient to produce 1.2 mg Phe 13Cδ-labelled or 0.8 mg Tyr 13Cδ-labelled purified ubiquitin with isotope labelling levels of 37% and 40%, respectively. In principle, using 3-13C pyruvate allows for a labelling degree of up to 50%, and we attribute the somewhat reduced isotope enrichment to a residual reservoir of unlabelled amino acids present in the eCells. We attempted to increase the level of isotope labelling by extensive dialysis of the eCells in phosphate buffered saline prior to use in CFPS, but this did not significantly increase the level of labelling. Most importantly, the presence of unlabelled Tyr or Phe did not affect the selectivity of labelling.
Selective 13C-labelling of the δ positions of phenylalanine or tyrosine in ubiquitin produced by eCell CFPS with 3-13C pyruvate as the carbon source. The plots show selected spectral regions from [13C,1H]-HSQC spectra using different labelling strategies. A CδH cross-peaks of the two phenylalanine residues of ubiquitin. The protein was produced in the presence of 3.5 mM unlabelled amino acids, including Tyr but not Phe. Protein yield was 1.2 mg from 300 mg eCells in 10 mL CFPS buffer. B CδH cross-peak of the single tyrosine residue of ubiquitin. The protein was produced in the presence of 3.5 mM unlabelled amino acids, including Phe but not Tyr. Protein yield 0.8 mg. C Uniformly 13C-labelled sample prepared using uniformly 13C-labelled glucose in a 5 mL eCell CFPS reaction containing 300 mg eCells, omitting Tyr and Phe. The yield was 1.7 mg purified protein
To demonstrate the general applicability of the labelling protocol, a sample of PpiB was made using 33 mM 3-13C pyruvate and 1.5 g eCells. 4.8 mg PpiB was obtained with a labelling level of about 36%. To increase the labelling degree of the Cδ positions, we also prepared a sample using 16 mM 1-13C glucose and 16.5 mM 3-13C pyruvate. The experiment yielded 2.7 mg protein per gram of eCells with about 50% 13C labelling of the Cδ positions. The [13C,1H]-spectra of the Cδ-labelled preparations showed resolved cross-peaks for 10 Phe and 3 Tyr residues free of multiplet splittings due to 1JCC couplings (Figures S1 and S2). PpiB contains 12 Phe and 3 Tyr residues, but some of them undergo ring flips in the intermediate time regime, where signals are broadened beyond detection (Takeda et al. 2010).
The biosynthetic pathways indicate that the ε carbons of Phe and Tyr and the δ2-carbon of Trp can be labelled also by using a different isotopologue precursor, 2-13C glucose. Using 1.5 g eCells in 25 mL CFPS buffer containing 30 mM 2-13C glucose yielded 4.8 mg PpiB with > 45% labelling degree of the ε carbons of Phe and Tyr. Figure 3 shows that this approach likewise avoided splittings by 1JCC couplings.
Selected spectral regions of the [1H,13C]-HSQC spectrum of PpiB produced by eCell CFPS with 2-13C glucose as the carbon source and with tyrosine, phenylalanine and tryptophan omitted and all other amino acids at 3.5 mM. The protein yield was 4.8 mg and the isotope labelling level about 45%. A CεH cross-peaks of tyrosine and CδH cross-peak of tryptophan. The weak unmarked cross-peak may arise from the CδH group of His8, as the production of histidine from 2-13C glucose directs 13C to the δ position of the imidazole ring (Weininger 2017). B CεH cross-peaks of phenylalanine. The two weak unmarked cross-peaks may arise from the CεH groups of Phe27 and Phe123, for which no assignments have been reported
While 3-13C pyruvate enriches only one of two Cδ positions in the aromatic rings of Phe and Tyr, the biosynthetic pathways (Fig. 1) suggest that the additional provision of 4-13C erythrose will label both Cδ positions and thus increase the sensitivity of the NMR measurements. Using 1.5 g eCells in 25 mL buffer containing 33 mM 3-13C pyruvate and 30 mM 4-13C erythrose, 1.6 mg PpiB were obtained with a labelling level of 70%. The spectrum showed singlets in the 13C dimension as expected (Fig. 4).
Discussion
Due to their relatively rigid and hydrophobic character, aromatic residues tend to be overrepresented in ligand binding sites and hydrophobic cores of proteins (Makwana and Mahalakshmi 2015). In enzymes, aromatic amino acids frequently form important interactions with substrates (Lanzarotti et al. 2011). This makes them attractive probes to assess 3D structure and ligand binding in pharmaceutical development and drug design (Brylinski 2018). To render aromatic side chains more amenable to monitoring by NMR spectroscopy, the spectral simplification afforded by atom-specific labelling confers an important benefit.
In contrast to protein production in vivo, where isotopic scrambling is best suppressed by using late metabolic precursors in isotope-labelled form together with auxotrophic bacterial strains, eCells provide a straightforward alternative route to atom-specific isotope labelling of aromatic amino acids starting from inexpensive and more readily available precursors. For example, the cost of isotope-labelled starting material for 4.8 mg PpiB produced from eCells was about 32 USD for 50 mg 2-13C glucose (Table 1). The equivalent sample produced in vivo would require much more isotope-labelled compound to satisfy the larger culture volumes involved. Typical amounts of 13C-labelled precursors used for expression in 1 L M9 minimal medium are 2.5 g 13C glucose, 2.0 g pyruvate or 1.3 g erythrose (Cai et al. 1998, 2019; Kasinath et al. 2013). The economics of eCell CFPS thus appear competitive to in vivo protein expression in minimal media also when measured in terms of cost of 13C-labelled precursor required per mg of isotope-labelled protein. We anticipate that actual outcomes will depend mostly on the intrinsic yields of the specific protein in eCell CFPS versus in vivo cell cultures.
Like in unlabelling experiments conducted in vivo (Rowlinson et al. 2022), provision of unlabelled amino acids in eCell CFPS allows suppressing undesired cross-peaks of the unlabelled amino-acid residues. This approach works well for discriminating between Phe and Tyr, as the E. coli metabolism does not easily swap carbon atoms between these two residue types (Takeuchi et al. 2007). The presence of tyrosine has been shown to differentially inhibit prephenate dehydrogenase (PDH; Hudson et al. 1983), thereby preventing 4-hydroxyphenyl pyruvic acid formation on the pathway to tyrosine (Fig. 1) and thus increasing the availability of the 13C-labelled precursors for the biosynthesis of other aromatic amino acids. Conversely, as PDH performs an irreversible catalytic step (Turnbull et al. 1990), unlabelled tyrosine is readily incorporated into the protein without diluting the pool of 13C-labelled intermediates. In a very similar way, prephenate dehydratase (PDT) is inhibited by as little as 50 µM phenylalanine (Gething et al. 1976). Finally, we found no evidence for eCell CFPS misdirecting 13C into non-targeted positions on the aromatic ring of the labelled amino acid. This establishes eCell CFPS as an attractive in vitro tool for selective labelling starting from inexpensive and readily accessible precursors.
The present work investigated the selective 13C enrichment of the δ or ε positions of aromatic amino acids by starting from different isotopologues. Like previous in vivo experiments conducted with selectively 13C-labelled glucose (Loquet et al. 2011), eCell CFPS achieves sparse 13C-labelling, where 13C enrichment can be targeted to the δ positions of Phe and Tyr residues using either the precursors 2-13C glucose (> 45%) or 3-13C pyruvate (> 36%). Even higher levels of 13C labelling in the δ position (> 70%) can be achieved by the combined use of 3-13C pyruvate and 4-13C erythrose. Previous efforts to label the δ positions of Phe and Tyr in vivo by the combined use of 1-13C glucose and 13C isotopologues of erythrose struggled to achieve labelling levels above 40% (Weininger 2017).
Some isotopic dilution due to a pool of unlabelled amino acids and precursors present in the eCells is difficult to avoid, as illustrated by our experiments conducted to label the δ positions of aromatic residues of PpiB by expression from 3-13C pyruvate and 1-13C glucose in eCells, where we obtained the expected labelling pattern (Figure S2) but a labelling level of only about 50%. Attempts to dialyse eCells in a large volume of buffer for an extended period to remove any unlabelled amino acids prior to eCell CFPS led to reduced protein yields, which may be expected as eCells gradually leak biomacromolecules (Van Raad and Huber 2021). Notably, however, our estimate of the labelling degree is conservative, as we compared the cross-peak volumes observed for the aromatic C–H groups with the cross-peak of 13C-pyruvate added to the solution as an internal standard. In a protein of the molecular weight of PpiB (19 kDa), transverse relaxation during the INEPT periods of the 13C-HSQC spectra would have affected the protein signals more than those of the internal standard.
In vivo experiments conducted with deuterated pyruvate and 4-13C erythrose have been shown to endow the δ positions of Tyr and Phe with single 1H-13C pairs with a labelling degree of 67% while simultaneously installing deuterium in neighbouring positions (Kasinath et al. 2013), which is particularly beneficial for 13C-relaxation measurements (Dreydoppel et al. 2021). While we did not perform experiments with deuterated compounds, we expect that the same labelling pattern would be obtained by eCell CFPS using the same precursors. This expectation is based on the result that eCells maintain the activities of the full complement of enzymes required for the biosynthesis of aromatic amino acids and thus perform in ways closely similar to live E. coli. For the same reason we anticipate that synthetic, commercially available late-stage precursors will perform as well in eCell CFPS as in in vivo protein expression.
Conclusions
The eCell system allows the selective 13C-enrichment of aromatic groups from early precursors, including atom-selective labelling of ring positions without 1J(13C,13C) couplings. High levels of isotope incorporation can be achieved with little expense for isotope-labelled precursors. Facile control over the chemical environment in eCell CFPS enables the ‘unlabelling’ of specific residues by simple addition of the respective unlabelled amino acids. The preparation of eCells is straightforward, can easily be scaled up and makes costly dialysis membranes redundant. By preserving the activities of the enzymes required for multi-step anabolic amino-acid biosynthesis from inexpensive precursors, the eCell CFPS platform is highly attractive for selective isotope labelling of proteins for NMR spectroscopy.
References
Akke M, Weininger U (2023) NMR studies of aromatic ring flips to probe conformational fluctuations in proteins. J Phys Chem B 127:591–599
Apponyi MA, Ozawa K, Dixon NE, Otting G (2008) Cell-free protein synthesis for analysis by NMR spectroscopy. Methods Mol Biol 426:257–268
Brylinski M (2018) Aromatic interactions at the ligand–protein interface: implications for the development of docking scoring functions. Chem Biol Drug Des 91:380–390
Cai M, Huang Y, Sakaguchi K, Clore GM, Gronenborn AM, Craigie R (1998) An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. J Biomol NMR 11:97–102
Cai M, Huang Y, Craigie R, Clore GM (2019) A simple protocol for expression of isotope-labeled proteins in Escherichia coli grown in shaker flasks at high cell density. J Biomol NMR 73:743–748
Dreydoppel M, Lichtenecker RJ, Akke M, Weininger U (2021) ) 1H R1r relaxation dispersion experiments in aromatic side chains. J Biomol NMR 75:383–392
Gell DA, Kwan AH, Mackay JP (2017) NMR spectroscopy in the analysis of protein-protein interactions. In: Webb GA (ed) Modern magnetic resonance. Springer International Publishing, Cham, Switzerland, pp 1–34
Gething MJH, Davidson BE, Dopheide TAA (1976) Chorismate mutase/prephenate dehydratase from Escherichia coli K12. 1. The effect of NaCl and its use in a new purification involving affinity chromatography on Sepharosyl-phenylalanine. Eur J Biochem 71:317–325
Hudson GS, Howlett GJ, Davidson BE (1983) The binding of tyrosine and NAD+ to chorismate mutase/prephenate dehydrogenase from Escherichia coli K12 and the effects of these ligands on the activity and self-association of the enzyme. Analysis in terms of a model. J Biol Chem 258:3114–3120
Jewett MC, Swartz JR (2004) Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng 86:19–26
Kainosho M, Güntert P (2009) SAIL – stereo-array isotope labeling. Quart Rev Biophys 42:247–300
Kasinath V, Valentine KG, Wand AJ (2013) A 13C labeling strategy reveals a range of aromatic side chain motion in calmodulin. J Am Chem Soc 135:9560–9563
Kurauskas V, Schanda P, Sounier R (2017) Methyl-specific isotope labeling strategies for NMR studies of membrane proteins. Methods Mol Biol 1635:109–123
Lacabanne D, Meier BH, Böckmann A (2018) Selective labeling and unlabeling strategies in protein solid-state NMR spectroscopy. J Biomol NMR 71:141–150
Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG (2011) Aromatic-aromatic interactions in proteins: beyond the dimer. J Chem Inf Model 51:1623–1633
Lichtenecker RJ (2014) Synthesis of aromatic 13C/2H-α-ketoacid precursors to be used in selective phenylalanine and tyrosine protein labelling. Org Biomol Chem 12:7551–7560
Lichtenecker RJ, Weinhäupl K, Schmid W, Konrat R (2013) α-Ketoacids as precursors for phenylalanine and tyrosine labelling in cell-based protein overexpression. J Biomol NMR 57:327–331
Linser R, Gelev V, Hagn F, Arthanari H, Hyberts SG, Wagner G (2014) Selective methyl labeling of eukaryotic membrane proteins using cell-free expression. J Am Chem Soc 136:11308–11310
Loquet A, Lv G, Giller K, Becker S, Lange A (2011) 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J Am Chem Soc 133:4722–4725
Lundström P, Teilum K, Carstensen T, Bezsonova I, Wiesner S, Hansen DF, Religa TL, Akke M, Kay LE (2007) Fractional 13C enrichment of isolated carbons using [1-13C]- or [2-13C]-glucose facilitates the accurate measurement of dynamics at backbone Cα and side-chain methyl positions in proteins. J Biomol NMR 38:199–212
Makwana KM, Mahalakshmi R (2015) Implications of aromatic–aromatic interactions: from protein structures to peptide models. Prot Sci 24:1920–1933
Milbradt AG, Arthanari H, Takeuchi K, Boeszoermenyi A, Hagn F, Wagner G (2015) Increased resolution of aromatic cross peaks using alternate 13C labeling and TROSY. J Biomol NMR 62:291–301
Pervushin K, Riek R, Wider G, Wüthrich K (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J Am Chem Soc 120:6394–6400
Rasia RM, Brutscher B, Plevin MJ (2012) Selective isotopic unlabeling of proteins using metabolic precursors: application to NMR assignment of intrinsically disordered proteins. ChemBioChem 13:732–739
Rowlinson B, Crublet E, Kerfah R, Plevin MJ (2022) Specific isotopic labelling and reverse labelling for protein NMR spectroscopy: using metabolic precursors in sample preparation. Biochem Soc Trans 50:1555–1567
Schörghuber J, Geist L, Platzer G, Feichtinger M, Bisaccia M, Scheibelberger L, Weber F, Konrat R, Lichtenecker RJ (2018) Late metabolic precursors for selective aromatic residue labeling. J Biomol NMR 71:129–140
Takeda M, Ono AM, Terauchi T, Miyano, Kainosho M (2010) Application of SAIL phenylalanine and tyrosine with alternative isotope-labeling patterns for protein structure determination. J Biomol NMR 46:45–49
Takeuchi K, Ng E, Malia TJ, Wagner G (2007) 1-13C amino acid selective labeling in a 2H/15 N background for NMR studies of large proteins. J Biomol NMR 38:89–98
Teilum K, Brath U, Lundström P, Akke M (2006) Biosynthetic 13C labeling of aromatic side chains in proteins for NMR relaxation measurements. J Am Chem Soc 128:2506–2507
Torizawa T, Shimizu M, Taoka M, Miyano H, Kainosho M (2004) Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J Biomol NMR 30:311–325
Torizawa T, Ono AM, Terauchi T, Kainosho M (2005) NMR assignment methods for the aromatic ring resonances of phenylalanine and tyrosine residues in proteins. J Am Chem Soc 127:12620–12626
Turnbull J, Cleland WW, Morrison JF (1990) Chorismate mutase-prephenate dehydrogenase from Escherichia coli. 1. Kinetic characterization of the dehydrogenase reaction by use of alternative substrates. Biochemistry 29:10245–10254
Van Raad D, Huber T (2021) In vitro protein synthesis in semipermeable artificial cells. ACS Synth Biol 10:1237–1244
Van Raad D, Otting G, Huber T (2023) Cell-free synthesis of proteins with selectively 13C-labelled methyl groups from inexpensive precursors. Magn Reson, in press. https://doi.org/10.5194/mr-2023-3
Verardi R, Traaseth NJ, Masterson LR, Vostrikov VV, Veglia G (2012) Isotope labeling for solution and solid-state NMR spectroscopy of membrane proteins. Adv Exp Med Biol 992:35–62
Weininger U (2017) Site-selective 13C labeling of proteins using erythrose. J Biomol NMR 67:191–200
Williams JK, Schmidt-Rohr K, Hong M (2015) Aromatic spectral editing techniques for magic-angle-spinning solid-state NMR spectroscopy of uniformly 13C-labeled proteins. Solid State Nucl Magn Reson 72:118–126
Acknowledgements
Financial support by the Australian Research Council (Grant No. FL170100019, DP200100348, DP21010088) and the Australian Research Council Centre of Excellence for Innovations in Peptide and Protein Science (Grant No. CE200100012) is gratefully acknowledged.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
DVR designed the experiments, wrote the first draft of the manuscript and prepared all figures, TH supervised the work, and GO and DVR wrote the final version of the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing financial interest(s): The Australian National University holds a patent related to this research (PCT/AU2020/050050) and share financial return from the patent with the inventors D. V. R. and T. H.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Van Raad, D., Huber, T. & Otting, G. Improved spectral resolution of [13C,1H]-HSQC spectra of aromatic amino acid residues in proteins produced by cell-free synthesis from inexpensive 13C-labelled precursors. J Biomol NMR 77, 183–190 (2023). https://doi.org/10.1007/s10858-023-00420-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-023-00420-9