Abstract
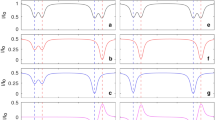
Protein molecules sample different conformations in solution and characterizing these conformations is crucial to understanding protein function. 15N CEST experiments are now routinely used to study slow conformational exchange of protein molecules between a ‘visible’ major state and ‘invisible’ minor states. These experiments have also been adapted to measure the solvent exchange rates of amide protons by exploiting the one bond deuterium isotope effect on the amide 15N chemical shifts. However at moderately high temperatures (~ 50 °C) that are sometimes required to populate protein minor conformers to levels (~ 1%) that can be detected by CEST experiments solvent H/D exchange can lead to ‘dips’ in low B1 15N CEST profiles that can be wrongly assigned to the conformational exchange process being characterized. This is demonstrated in the case of ~ 18 kDa T4 Lysozyme (T4L) at 50 °C and the ~ 11 kDa E. coli hibernation promoting factor (HPF) at 52 °C. This problem is trivially solved by eliminating the exchangeable deuterons in the solvent by using either an external D2O lock or by using a small amount (~ 1–3%) of a molecule like d6-DMSO that does not contain exchangeable deuterons to lock the spectrometer.


Similar content being viewed by others
References
Ahlner A, Carlsson M, Jonsson BH, Lundstrom P (2013) PINT: a software for integration of peak volumes and extraction of relaxation rates. J Biomol NMR 56:191–202
Arakawa T, Kita Y, Timasheff SN (2007) Protein precipitation and denaturation by dimethyl sulfoxide. Biophys Chem 131:62–70
Austin RH, Beeson KW, Eisenstein L, Frauenfelder H, Gunsalus IC (1975) Dynamics of ligand binding to myoglobin. Biochemistry 14:5355–5373
Bouvignies G (2012) Chemex (https://github.com/gbouvignies/chemex/releases)
Bouvignies G, Vallurupalli P, Kay LE (2014) Visualizing side chains of invisible protein conformers by solution NMR. J Mol Biol 426:763–774
Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ (2006) Protein NMR spectroscopy, principles and practice, 2nd edn. Academic Press, Cambridge
Ceccon A, Clore GM, Tugarinov V (2018) Decorrelating kinetic and relaxation parameters in exchange saturation transfer NMR: a case study of N-terminal huntingtin peptides binding to unilamellar lipid vesicles. J Phys Chem B 122:11271–11278
Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR (2010) 15NH/D-SOLEXSY experiment for accurate measurement of amide solvent exchange rates: application to denatured drkN SH3. J Biomol NMR 46:227–244
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe—a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293
Deshmukh L, Tugarinov V, Appella DH, Clore GM (2018) Targeting a dark excited state of HIV-1 nucleocapsid by antiretroviral thioesters revealed by NMR spectroscopy. Angew Chem 57:2687–2691
Eriksson AE, Baase WA, Wozniak JA, Matthews BW (1992) A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature 355:371–373
Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM (2011) Atomic-resolution dynamics on the surface of amyloid-beta protofibrils probed by solution NMR. Nature 480:268–272
Forsen S, Hoffman RA (1963) Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J Chem Phys 39:2892–2901
Gladkova C, Schubert AF, Wagstaff JL, Pruneda JN, Freund SMV, Komander D (2017) An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J 36:3555–3572
Goddard TD, Kneller DG (2008) SPARKY 3 University of California, San Francisco
Gopalan AB, Vallurupalli P (2018) Measuring the signs of the methyl 1H chemical shift diferences between major and ‘invisible’ minor protein conformational states using methyl 1H multi-quantum spectroscopy. J Biomol NMR 70:187–202
Guenneugues M, Berthault P, Desvaux H (1999) A method for determining B1 field inhomogeneity. Are the biases assumed in heteronuclear relaxation experiments usually underestimated? J Magn Reson 136:118–126
Gupta RK, Redfield AG (1970) Double nuclear magnetic resonance observation of electron exchange between ferri- and ferrocytochrome c. Science 169:1204–1206
Hansen PE (2000) Isotope effects on chemical shifts of proteins and peptides. Magn Reson Chem 38:1–10
Hansen AL, Kay LE (2014) Measurement of histidine pKa values and tautomer populations in invisible protein states. Proc Natl Acad Sci USA 111:E1705–E1712
Hansen DF, Led JJ (2006) Determination of the geometric structure of the metal site in a blue copper protein by paramagnetic NMR. Proc Natl Acad Sci USA 103:1738–1743
Hansen AL, Bouvignies G, Kay LE (2013) Probing slowly exchanging protein systems via 13Cα-CEST: monitoring folding of the Im7 protein. J Biomol NMR 55:279–289
Karplus M (2000) Aspects of protein reaction dynamics: deviations from simple behavior. J Phys Chem B 104:11–27
Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE (2004) Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430:586–590
Kumari P, Frey L, Sobol A, Lakomek NA, Riek R (2018) 15N transverse relaxation measurements for the characterization of micros-ms dynamics are deteriorated by the deuterium isotope effect on (15)N resulting from solvent exchange. J Biomol NMR 72(3–4):125–137
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327
Leninger M, Marsiglia WM, Jerschow A, Traaseth NJ (2018) Multiple frequency saturation pulses reduce CEST acquisition time for quantifying conformational exchange in biomolecules. J Biomol NMR 71:19–30
Lim J, Xiao TS, Fan JS, Yang DW (2014) An off-pathway folding intermediate of an acyl carrier protein domain coexists with the folded and unfolded states under native conditions. Angew Chem Int Edit 53:2358–2361
Liu LJ, Baase WA, Matthews BW (2009) Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I–S and I–Se halogen-bonding. J Mol Biol 385:595–605
Long D, Bouvignies G, Kay LE (2014) Measuring hydrogen exchange rates in invisible protein excited states. Proc Natl Acad Sci USA 111:8820–8825
Maki Y, Yoshida H, Wada A (2000) Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965–974
McConnell HM (1958) Reaction rates by nuclear magnetic resonance. J Chem Phys 28:430–431
Milojevic J, Esposito V, Das R, Melacini G (2007) Understanding the molecular basis for the inhibition of the Alzheimer’s Aβ-peptide oligomerization by human serum albumin using saturation transfer difference and off-resonance relaxation NMR spectroscopy. J Am Chem Soc 129:4282–4290
Morris GA, Freeman R (1978) Selective excitation in Fourier-transform nuclear magnetic-resonance. J Magn Reson 29:433–462
Palmer AG III, Kroenke CD, Loria JP (2001) Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol 339:204–238
Palmer AG III (2014) Chemical exchange in biomacromolecules: past, present, and future. J Magn Reson 241:3–17
Palmer AG, Massi F (2006) Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem Rev 106:1700–1719
Sato A, Watanabe T, Maki Y, Ueta M, Yoshida H, Ito Y, Wada A, Mishima M (2009) Solution structure of the E. coli ribosome hibernation promoting factor HPF: implications for the relationship between structure and function. Biochem Biophys Res Commun 389:580–585
Sekhar A, Kay LE (2013) NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci USA 110:12867–12874
Sekhar A, Velyvis A, Zoltsman G, Rosenzweig R, Bouvignies G, Kay LE (2018) Conserved conformational selection mechanism of Hsp70 chaperone-substrate interactions. Elife 7:e32764
Thakur A, Chandra K, Dubey A, D’Silva P, Atreya HS (2013) Rapid characterization of hydrogen exchange in proteins. Angew Chem Int Ed Engl 52:2440–2443
Vallurupalli P, Hansen DF, Lundstrom P, Kay LE (2009) CPMG relaxation dispersion NMR experiments measuring glycine 1H alpha and 13C alpha chemical shifts in the ‘invisible’ excited states of proteins. J Biomol NMR 45:45–55
Vallurupalli P, Bouvignies G, Kay LE (2012) Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc 134:8148–8161
Vallurupalli P, Sekhar A, Yuwen T, Kay LE (2017) Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: a primer. J Biomol NMR 67:243–271
Ward KM, Aletras AH, Balaban RS (2000) A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson 143:79–87
Yuwen T, Sekhar A, Kay LE (2017) Separating dipolar and chemical exchange magnetization transfer processes in 1H-CEST. Angew Chem Int Ed Engl 56:6122–6125
Yuwen T, Bah A, Brady JP, Ferrage F, Bouvignies G, Kay LE (2018a) Measuring solvent hydrogen exchange rates by multifrequency excitation 15N CEST: application to protein phase separation. J Phys Chem B 122(49):11206–11217
Yuwen T, Bouvignies G, Kay LE (2018b) Exploring methods to expedite the recording of CEST datasets using selective pulse excitation. J Magn Reson 292:1–7
Yuwen T, Kay LE, Bouvignies G (2018c) Dramatic Decrease in CEST Measurement Times Using Multi-Site Excitation. Chemphyschem 19:1707–1710
Zaiss M, Bachert P (2013) Chemical exchange saturation transfer (CEST) and MR Z-spectroscopy in vivo: a review of theoretical approaches and methods. Phys Med Biol 58:R221–R269
Zhao B, Hansen AL, Zhang Q (2014) Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R1ρ NMR spectroscopy. J Am Chem Soc 136:20–23
Acknowledgements
We would like to thank Dr. Tairan Yuwen and Prof. Lewis Kay (University of Toronto) for providing the Bruker 15N CEST pulse sequence, Dr. G Bouvignies for making ChemEx along with the source code freely available, the national NMR facility at TIFR, Hyderabad and Dr. Krishna Rao for spectrometer time. The work was supported by generous funds from TCIS/TIFRH and Grant No. ECR/2016/001088 from SERB awarded to PV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tiwari, V.P., Pandit, S. & Vallurupalli, P. Exchangeable deuterons introduce artifacts in amide 15N CEST experiments used to study protein conformational exchange. J Biomol NMR 73, 43–48 (2019). https://doi.org/10.1007/s10858-018-00223-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-018-00223-3