Abstract
Anionic complex of antamanide with the nitrate anion has been proven by electrospray ionization mass spectrometry (ESI-MS) method. Further, applying quantum chemical DFT calculations, the most probable structure of this complex was derived. The nitrate anion is embedded in the molecule of antamanide and its oxygens atoms are bonded by seven bonds to the hydrogen atoms of the ligand. Finally, the interaction energy, E(int), of the antamanide-NO3− complex was calculated as E(int) = -175.9 kJ/mol.
Highlights
-
Antamanide-NO3− complex has been proved using the ESI-MS method.
-
The structures of this complex has been determined applying quantum chemical DFT calculations.
-
The interaction energies of antamanide-NO3− complex has been calculated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cyclic decapeptide antamanide, cycl[-Val(1)-Pro(2) -Pro(3)-Ala(4)-Phe(5)-Phe(6)-Pro(7)-Pro(8)-Phe(9)-Phe(10)-], C64H78N10O10, see Scheme 1, consists entirely of l-amino acids. Antamanide was isolated from the poisonous mushroom Amanita phalloides and it has the unique property of counteracting the toxin phalloidin, produced by the mentioned mushroom. Furthermore, the antamanide was also prepared by synthetic way [1, 2]. Moreover, it should be noted that various conformations of this electroneutral decapeptide compound have been reported [3,4,5].
Cyclic peptides are also somewhat more “rigid” compared to the corresponding linear peptides, and this attribute promotes binding by removing the “entropic penalty” [5]. Falvo et al. found strong intramolecular hydrogen bonding network in the molecule of antamanide [4].
Antamanide forms 1:1 complexes with a variety of metal cations [6,7,8,9]. In our previous works, we dealt with the complexes of antamanide with univalent [10,11,12,13,14] and divalent cations [15,16,17]. The structures of antamanide complexes have been studied by the quantum mechanical DFT calculations. It was found that the cations are embedded in the cavity of antamanide. Univalent cations H+, K+, Tl+ and H3O+ are bonded by four bonds to four oxygens of antamanide molecule, while the divalent Ca2+, Sr2+ and Ba2+ are bonded by six bonds to the six antamanide oxygens.
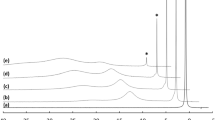
Mass spectrum in the negative ESI mode of a mixture of antamanide (5*10− 6 mol/L) and TBA+NO3− (2.5*10− 4 mol/L) in chloroform/methanol, 1:1 (ν/ν). Anionic complex of antamanide with NO3− in the ratio 1/1, [C64H78N10O10 + NO3]−, provides ion cluster at m/z 1208.6. The insets show (a) the zoomed view of the measured ion cluster and (b) the isotope model of the ion cluster of the complex, respectively. Very intense ion cluster at m/z 366.3 belongs to complex of TBA+ with two nitrate anions
Zhu et al. [18] and Becherer et al. [19] investigated the formation of anionic complexes of various macrocycles. Reviews of the complex formation of macrocycles with anions published Kubik [20] and Elmes and Jolliffe [21]. The latter authors pointed out that the large peptides proteins give the possibility of numerous H-bonding interactions available from the different amino acids that are present in the peptide molecule. Kubik reported macrocyclic complexes of halides, nitrate, hydrogensulfate, and dihydrogenphosphate anions [20].
However, up to now, the investigation of the antamanide H-bonding with the anions has not been made. So, in the current work, electrospray ionization mass spectrometry (ESI-MS) was used for the investigation of the interaction between the univalent nitrate anion and the antamanide ligand in the gas phase. Further, employing quantum mechanical DFT calculations, the most probable structure of this experimentally proven anionic complex in the gas phase was predicted.
Experimental
Antamanide was supplied by the Padua Unit of the IBCCNR, Italy. Other chemicals used (Lachema, Brno, Czech Republic) were of reagent grade purity.
The experiments were provided with a 3200 Q TRAP (AB Sciex, Canada) mass spectrometer fitted with an electrospray ion source. Antamanide and tetrabutylammonium nitrate (TBA+NO3−) were dissolved in chloroform/methanol, 1:1 (ν/ν). Final concentration of antamanide was 5*10− 6 mol/L and that of TBA+NO3− 2.5*10− 4 mol/L. The dissolved mixture was introduced into the ESI source via a PEEK capillary at a flow rate of 10 µL/min. Nitrogen was used as a nebulizer and collision gas. The operating conditions for the mass spectrometer were set as follows: ionspray voltage − 4.5 kV; curtain gas 10, ion source gas(1) 18, and ion source gas(2) 0 psig; ion source temperature ambient; declustering potential − 60 V, entrance potential − 10 V.
The theoretical calculations have been performed in the frame of the density functional theory with hybrid density functional, “Becke, 3-parameter, Lee-Yang-Parr” (B3LYP) version [22]. Gaussian 16 set of programs was used for all calculations [23]. In order to stabilize self-consistent field procedure, the scaled steepest descent algorithm was used during optimization. The optimization itself was unconstrained. LanL2DZ (Los Alamos National Laboratory 2 double ζ) basis set [24, 25] was used for the respective calculations.
The interaction energies between antamanide and the nitrate anion was calculated with basis set super position error correction. The counterpoise method, as it is implemented in Gaussian 16 package program, was used. The convergence energy criteria were set to E < 10− 9 hartree.
The Gaussian keyword “integral(ultrafinegrid)” was used in order to increase the numerical accuracy and to reduce oscillations during the molecular geometry optimization. So, two-electron integrals and their derivatives were calculated by using the pruned (99,590) integration grid, having 99 radial shells and 590 angular points per shell.
Results and discussion
Figure 1 shows an ESI spectrum obtained in negative mode for a mixture of antamanide with TBA+NO3- in chloroform/methanol mixture (1:1, ν/ ν). The ion cluster at m/z 1208.6 belongs to the anionic complex of antamanide with nitrate anion, [C64H78N10O10 + NO3]-. Very intense ion cluster at m/z 366.3 belongs to complex of TBA+ with two nitrate anions. The insets in Fig. 1 represent the zoomed view of the measured ion cluster and the appropriate model spectrum, respectively. As can be seen the measured ion cluster and a theoretical one are very close. The heights of the peaks correspond to the content of 13 C in natural carbon (1.1%) and to 64 carbon atoms in the molecule of antamanide.
The existence of the above mentioned 1/1 complex of antamanide with NO3- was further proofed by a tandem mass spectrometry experiment.
Figure 2 shows enhanced product ion spectrum of the precursor m/z 1208.6, [C64H78N10O10 + NO3]-. In the spectrum, it is possible to distinguish a signal related to the presence of the original precursor. The other signal obtained by collision induced dissociation (CID) relates to a loss of HNO3 from the precursor leading to the fragment possessing the structure [C64H78N10O10-H]-.
So, the anionic complex of antamanide with one nitrate anion has been proved by mass spectrometry.
The most probable conformations of the antamanide – NO3- complex has been determined based on the thorough conformational analysis (i.e., six very different initial mutual positions of the ligand and the nitrate anion were considered during the geometry optimization) and the respective vibrational frequency calculations, analogously as in our previous articles [11,12,13,14,15,16,17]. The molecular geometries of the free ligand and its complex with NO3- were optimized. The optimized conformation of this free ligand is given in Fig. 3.
In Fig. 4, the structure obtained by the full DFT optimization of the antamanide-NO3- complex is depicted, together with the lengths of the corresponding bonds (in Å). In the resulting anionic complex species, which is most energetically favored, the three oxygens of the “central” anion NO3- is bound by seven hydrogen bonds to the respective seven hydrogen atoms of the parent antamanide ligand.
The interaction between the atoms of nitrate anion and others closest atoms is described in the frame of the Wiberg bond index [26] after deduction of mutual bonds in NO3- (Table 1).
Furthermore, the interaction energy, E(int), of the antamanide-NO3- complex [calculated as the respective difference between the pure electronic energies of the respective complex and isolated antamanide and NO3- species: E(int) = [E(complex) – E(antamanide) – E(NO3-)] was calculated. Energies were calculated in optimized geometry from B3LYP/LanL2DZ approach. For these calculations, we used B3LYP functional with LanL2DZ resp. Def2TZVP basis sets and M06 functional with Def2TZVP basis sets, with and without the dispersion correction [27,28,29,30,31]. The results are summarized in Table 2.
It is clear from Table 2 that the calculated interaction energies for the B3LYP functional are almost the same despite the basis set used, while the M06 functional gives slightly higher interaction energy values. The introduction of the dispersion correction always leads to a little bit higher values of the calculated interaction energy.
Conclusion
In this work, we have shown that the combination of theoretical DFT calculations and an experimental ESI-MS method can provide relevant data on the noncovalent bonding interactions of the nitrate anion NO3− with the antamanide ligand. By using this experimental method, the anionic antamanide – NO3− complex has been evidenced quite unambiguously. An anionic complex of antamanide has been here described for the first time. Applying DFT calculations, the most probable structure of this complex was derived. In this anionic complex, the NO3− anion is embedded in the molecule of antamanide. Two or three bonds to the hydrogen atoms of antamanide molecule bond each oxygen atom of this anion. The interaction energy, E(int), of the antamanide-NO3− complex has been calculated as -175.9 kJ/mol.
References
Wieland, T., Lüben, G., Ottenheym, H., Faesel, J., de Vries, J.X., Prox, A., Schmid, J.: The Discovery, isolation, elucidation of structure, and synthesis of Antamanide. Angew Chem. Int. Ed. Engl. 7, 204–208 (1968). https://doi.org/10.1002/anie.196802041
Wieland, T., Faulstich, H., Fiume, L.: Amatoxins, Phallotoxins, Phallolysin, and Antamanide: The biologically active components of poisonous Amanita mushroom. Crit. Rev. Biochem. 5, 185–260 (1978). https://doi.org/10.3109/10409237809149870
Moran, A.M., Park, S.M., Mukamel, S.: Infrared photon echo signatures of hydrogen bond connectivity in the cyclic decapeptide antamanide. J. Chem. Phys. 118, 9971–9980 (2003). https://doi.org/10.1063/1.1571527
Falvo, C., Hayashi, T., Zhuang, W., Mukamel, S.: Coherent two dimensional infrared spectroscopy of a cyclic decapeptide antamanide. A simulation study of the amide-I and a bands. J. Phys. Chem. B. 112, 12479–12490 (2008). https://doi.org/10.1021/jp801493y
Craik, D.J.: Seamless proteins tie up their Loose ends. Science. 311, 1563–1564 (2006). https://doi.org/10.1126/science.1125248
Wieland, T., Faulstich, H., Burgemeister, W.: Antamanide and analogs. Studies on selectivity and stability of complexes. Biochem. Biophys. Res. Commun. 47, 984–992 (1972). https://doi.org/10.1016/0006-291X(72)90929-1
Wieland, T., Faulstich, H., Burgemeister, W., Otting, W., Möhle, W., Shemyakin, M.M., Ovchinnikov, Y.A., Ivanov, V.T., Malenkov, G.G.: Affinity of antamanide for sodium ions. FEBS Lett. 9, 89–92 (1970). https://doi.org/10.1016/0014-5793(70)80320-9
Pangavhane, S., Makrlík, E., Ruzza, P., Kašička, V.: Affinity capillary electrophoresis employed for determination of stability constants of antamanide complexes with univalent and divalent cations in methanol. Electrophoresis. 40, 2321–2328 (2019). https://doi.org/10.1002/elps.201900043
Ruzza, P., Calderan, A., Biondi, B., Carrara, M., Tancredi, T., Borin, G.: Ion-binding and pharmacological properties of Tyr6 and Tyr9 antamanide analogs. J. Pept. Res. 53, 442–452 (1999). https://doi.org/10.1034/j.1399-3011.1999.00018.x
Makrlík, E., Vaňura, P., Ruzza, P.: Synergistic extraction of some Univalent cations from Water into Nitrobenzene using Sodium Dicarbollylcobaltate and Antamanide. J. Solut. Chem. 46, 1121–1130 (2017). https://doi.org/10.1007/s10953-017-0625-8
Makrlík, E., Böhm, S., Vaňura, P., Ruzza, P.: Protonation of antamanide: Experimental and theoretical study. J. Mol. Liq. 196, 163–166 (2014). https://doi.org/10.1016/j.molliq.2014.03.019
Makrlík, E., Böhm, S., Vaňura, P., Ruzza, P.: Complexation of Li + with antamanide: An experimental and theoretical study. Monatsh Chem. 145, 1051–1054 (2014). https://doi.org/10.1007/s00706-014-1165-1
Makrlík, E., Vaňura, P., Böhm, S., Ruzza, P.: Experimental and theoretical study on interaction of the potassium cation with antamanide. Chem. Phys. 433, 85–88 (2014). https://doi.org/10.1016/j.chemphys.2014.02.002
Makrlík, E., Böhm, S., Vaňura, P., Ruzza, P.: Interaction of the univalent thallium cation with antamanide: Experimental and theoretical study. J. Mol. Struct. 1064, 107–110 (2014). https://doi.org/10.1016/j.molstruc.2014.01.089
Makrlík, E., Böhm, S., Vaňura, P., Ruzza, P.: Complexation of the calcium cation with antamanide: An experimental and theoretical study. Mol. Phys. 113, 1472–1477 (2015). https://doi.org/10.1080/00268976.2015.1006276
Makrlík, E., Vaňura, P., Böhm, S., Ruzza, P.: Extraction and theoretical study on complexation of the strontium cation with antamanide. J. Radioanal Nucl. Chem. 300, 1291–1294 (2014). https://doi.org/10.1007/s10967-014-3098-x
Makrlík, E., Böhm, S., Vaňura, P., Ruzza, P.: Experimental and theoretical study on interaction of the barium cation with antamanide. J. Mol. Struct. 1065–1066, 61–64 (2014). https://doi.org/10.1016/j.molstruc.2014.02.009
Zhu, S.S., Staats, H., Brandhorst, K., Grunenberg, J., Gruppi, F., Dalcanale, E., Lutzen, A., Rissanen, K., Schalley, C.A.: Anion binding to Resorcinarene-Based Cavitands: The importance of C-H+···Anion interactions. Angew Chem. Int. Ed. 47, 788–792 (2008). https://doi.org/10.1002/anie.200703451
Becherer, T., Meshcheryakov, D., Springer, A., Bohmer, V., Schalleya, C.A.: Mass spectrometric study of oligourea macrocycles and their anion binding behavior. J. Mass. Spectrom. 44, 1338–1347 (2009). https://doi.org/10.1002/jms.1616
Kubik, S.: Amino acid containing anion receptors. Chem. Soc. Rev. 38, 585–605 (2009). https://doi.org/10.1039/b810531f
Elmes, R.B.P., Jolliffe, K.A.: Anion recognition by cyclic peptides. Chem. Commun. 51, 4951–4968 (2015). https://doi.org/10.1039/c4cc10095f
Lee Chengteh, Sosa, C.: Local density component of the Lee–Yang–Parr correlation energy functional. J. Chem. Phys. 100, 9018–9024 (1994). https://doi.org/10.1063/1.466706
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Petersson, G.A., Nakatsuji, H., Li, X., Caricato, M., Marenich, A.V., Bloino, J., Janesko, B.G., Gomperts, R., Mennucci, B., Hratchian, H.P., Ortiz, J.V., Izmaylov, A.F., Sonnenberg, J.L., Williams-Young, D., Ding, F., Lipparini, F., Egidi, F., Goings, J., Peng, B., Petrone, A., Henderson, T., Ranasinghe, D., Zakrzewski, V.G., Gao, J., Rega, N., Zheng, G., Liang, W., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Throssell, K., Montgomery, J.A. Jr., Peralta, J.E., Ogliaro, F., Bearpark, M.J., Heyd, J.J., Brothers, E.N., Kudin, K.N., Staroverov, V.N., Keith, T.A., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A.P., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Millam, J.M., Klene, M., Adamo, C., Cammi, R.: Ochterski J. W., Martin R. L., Morokuma K., Farkas O., Foresman J. B., Fox D. J. Gaussian, Inc., Wallingford CT, 2016.
Hay, P.J., Wadt, W.R.: Ab initio effective core potentials for molecular calculations – potentials for the transition-metal atoms sc to hg. J. Chem. Phys. 82, 270–283 (1985). https://doi.org/10.1063/1.448799
Wadt, W.R., Hay, P.J.: Ab initio effective core potentials for molecular calculations – potentials for main group elements na to Bi. J. Chem. Phys. 82, 284–298 (1985). https://doi.org/10.1063/1.448800
Wiberg, K.B.: Application of the pople-santry-segal CNDO method to the cyclopropylcarbinyl and cyclobutyl cation and to bicyclobutane. Tetrahedron. 24, 1083–1096 (1968). https://doi.org/10.1016/0040-4020(68)88057-3
Dykstra, C.E.: Theory and Applications of Computational Chemistry, 1st edn. Elsevier, Amsterdam (2005)
Liang, G., de Yonker, N.J., Zhao, X., Webster, C.E.: Prediction of the reduction potential in transition-metal containing complexes: How expensive? For what accuracy. J. Comput. Chem. 38, 2430–2438 (2017). https://doi.org/10.1002/jcc.24894
Qi, X.-J., Fu, Y., Liu, L., Guo, Q.-X.: Ab Initio Calculations of Thermodynamic Hydricities of Transition-Metal Hydrides in Acetonitrile. Organometallics. 26, 4197–4203 (2007). https://doi.org/10.1021/om0702429
Wanno, B., Tabtimsai, C.: A DFT investigation of CO adsorption on VIIIB transition metal-doped graphene sheets. Superlatt Microstruct. 67, 110–117 (2014). https://doi.org/10.1016/j.spmi.2013.12.025
Binbay, N.: Comparison of LANL2DZ and SBKJC basis sets, over DFT calculations of a typical half-sandwich ruthenium complex. Russ J. Phys. Chem. 96, 3161–3169 (2022). https://doi.org/10.1134/S0036024423020188
Acknowledgements
The Czech Ministry of Education, Youth, and Sports, Project MSMT No., supported this work: 20/2015. Computational resources were provided by the e-INFRA CZ project (ID:90140), supported by the Ministry of Education, Youth and Sports of the Czech Republic.
Funding
Open access publishing supported by the National Technical Library in Prague.
Author information
Authors and Affiliations
Contributions
Author contributions PV wrote the most of the manuscript text, DS measured the MS spectra, TU provided DFT calculations. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vaňura, P., Sýkora, D. & Uhlíková, T. Complex of antamanide with the nitrate anion. J Incl Phenom Macrocycl Chem 103, 385–391 (2023). https://doi.org/10.1007/s10847-023-01199-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10847-023-01199-w