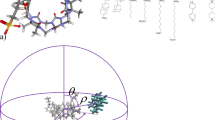
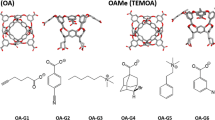
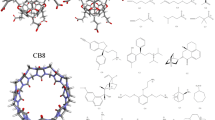
Abstract
Statistical Assessment of Modeling of Proteins and Ligands (SAMPL) challenges provide routes to compare chemical quantities determined using computational chemistry approaches to experimental measurements that are shared after the competition. For this effort, several computational methods have been used to calculate the binding energies of Octa Acid (OA) and exo-Octa Acid (exoOA) host–guest systems for SAMPL7. The initial poses for molecular dynamics (MD) were generated by molecular docking. Binding free energy calculations were performed using molecular mechanics combined with Poisson–Boltzmann or generalized Born surface area solvation (MMPBSA/MMGBSA) approaches. The factors that affect the utility of the MMPBSA/MMGBSA approaches including solvation, partial charge, and solute entropy models were also analyzed. In addition to MD calculations, quantum mechanics (QM) calculations were performed using several different density functional theory (DFT) approaches. From SAMPL6 results, B3PW91-D3 was found to overestimate binding energies though it was effective for geometry optimizations, so it was considered for the DFT geometry optimizations in the current study, with single-point energy calculations carried out with B2PLYP-D3 with double-, triple-, and quadruple-ζ level basis sets. Accounting for dispersion effects, and solvation models was deemed essential for the predictions. MMGBSA and MMPBSA correlated better to experiment when used in conjunction with an empirical/linear correction.







Similar content being viewed by others
References
Alonso H, Bliznyuk AA, Gready JE (2006) Combining docking and molecular dynamic simulations in drug design. Med Res Rev 26:531–568. https://doi.org/10.1002/med.20067
Brown FK, Sherer EC, Johnson SA et al (2017) The evolution of drug design at Merck Research Laboratories. J Comput Aided Mol Des 31:255–266. https://doi.org/10.1007/s10822-016-9993-1
Cerchietti LC, Ghetu AF, Zhu X et al (2010) A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17:400–411. https://doi.org/10.1016/j.ccr.2009.12.050
Jiang X, Dulubova I, Reisman SA et al (2020) A novel series of cysteine-dependent, allosteric inverse agonists of the nuclear receptor RORγt. Bioorg Med Chem Lett 30:126967. https://doi.org/10.1016/j.bmcl.2020.126967
Muddana HS, Daniel Varnado C, Bielawski CW et al (2012) Blind prediction of host–guest binding affinities: a new SAMPL3 challenge. J Comput Aided Mol Des 26:475–487. https://doi.org/10.1007/s10822-012-9554-1
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) The SAMPL4 host–guest blind prediction challenge: an overview. J Comput Aided Mol Des 28:305–317. https://doi.org/10.1007/s10822-014-9735-1
Yin J, Henriksen NM, Slochower DR et al (2017) Overview of the SAMPL5 host–guest challenge: Are we doing better? J Comput Aided Mol Des 31:1–19. https://doi.org/10.1007/s10822-016-9974-4
Nicholls A, Wlodek S, Grant JA (2009) The SAMP1 solvation challenge: Further lessons regarding the pitfalls of parametrization. J Phys Chem B 113:4521–4532. https://doi.org/10.1021/jp806855q
Geballe MT, Skillman AG, Nicholls A et al (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24:259–279. https://doi.org/10.1007/s10822-010-9350-8
Rizzi A, Murkli S, McNeill JN et al (2018) Overview of the SAMPL6 host-guest binding affinity prediction challenge. J Comput Aided Mol Des 32(10):937–963
Işık M, Bergazin TD, Fox T et al (2020) Assessing the accuracy of octanol–water partition coefficient predictions in the SAMPL6 Part II log P Challenge. Springer, New York
Işık M, Levorse D, Rustenburg AS et al (2018) pK a measurements for the SAMPL6 prediction challenge for a set of kinase inhibitor-like fragments. J Comput Aided Mol Des 32:1117–1138. https://doi.org/10.1007/s10822-018-0168-0
Gibb CLD, Gibb BC (2014) Binding of cyclic carboxylates to octa-acid deep-cavity cavitand. J Comput Aided Mol Des 28:319–325. https://doi.org/10.1007/s10822-013-9690-2
Zwanzig RW (1954) High-temperature equation of state by a perturbation method. I Nonpolar gases. J Chem Phys 22:1420–1426. https://doi.org/10.1063/1.1740409
Jiang W, Hodoscek M, Roux B (2009) Computation of absolute hydration and binding free energy with free energy perturbation distributed replica-exchange molecular dynamics. J Chem Theory Comput 5:2583–2588. https://doi.org/10.1021/ct900223z
Mitchell MJ, McCammon JA (1991) Free energy difference calculations by thermodynamic integration: difficulties in obtaining a precise value. J Comput Chem 12:271–275. https://doi.org/10.1002/jcc.540120218
Miller BR, McGee TD, Swails JM et al (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314–3321. https://doi.org/10.1021/ct300418h
Procacci P, Guarrasi M, Guarnieri G (2018) SAMPL6 host–guest blind predictions using a non equilibrium alchemical approach. J Comput Aided Mol Des 32:965–982. https://doi.org/10.1007/s10822-018-0151-9
Frank L, Nupur S, Zheng Z, Merz KM (2018) Detailed potential of mean force studies on host–guest systems from the SAMPL6 challenge. J Comput Aided Mol Des 32:1013–1026. https://doi.org/10.1007/s10822-018-0153-7
Caldararu O, Olsson MA, Misini Ignjatović M et al (2018) Binding free energies in the SAMPL6 octa-acid host–guest challenge calculated with MM and QM methods. J Comput Aided Mol Des 32:1027–1046. https://doi.org/10.1007/s10822-018-0158-2
Nishikawa N, Han K, Wu X et al (2018) Comparison of the umbrella sampling and the double decoupling method in binding free energy predictions for SAMPL6 octa-acid host–guest challenges. J Comput Aided Mol Des 32:1075–1086. https://doi.org/10.1007/s10822-018-0166-2
Laury ML, Wang Z, Gordon AS, Ponder JW (2018) Absolute binding free energies for the SAMPL6 cucurbit[8]uril host–guest challenge via the AMOEBA polarizable force field. J Comput Aided Mol Des 32:1087–1095. https://doi.org/10.1007/s10822-018-0147-5
Eken Y, Patel P, Díaz T et al (2018) SAMPL6 host–guest challenge: binding free energies via a multistep approach. J Comput Aided Mol Des 32:1097–1115. https://doi.org/10.1007/s10822-018-0159-1
Dixon T, Lotz SD, Dickson A (2018) Predicting ligand binding affinity using on- and off-rates for the SAMPL6 SAMPLing challenge. J Comput Aided Mol Des 32:1001–1012. https://doi.org/10.1007/s10822-018-0149-3
Hudson PS, Han K, Woodcock HL, Brooks BR (2018) Force matching as a stepping stone to QM/MM CB[8] host/guest binding free energies: a SAMPL6 cautionary tale. J Comput Aided Mol Des 32:983–999. https://doi.org/10.1007/s10822-018-0165-3
Sun H, Duan L, Chen F et al (2018) Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys Chem Chem Phys 20:14450–14460. https://doi.org/10.1039/c7cp07623a
Hamaguchi N, Fusti-Molnar L, Wlodek S (2012) Force-field and quantum-mechanical binding study of selected SAMPL3 host-guest complexes. J Comput Aided Mol Des 26:577–582. https://doi.org/10.1007/s10822-012-9553-2
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652. https://doi.org/10.1063/1.464913
Lee C, Yang E, Parr RG (1988) Development of the Colic-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37(2):785
Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ (1994) Ab Initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J Phys Chem 98:11623–11627. https://doi.org/10.1021/j100096a001
Sure R, Antony J, Grimme S (2014) Blind prediction of binding affinities for charged supramolecular host-guest systems: achievements and shortcomings of DFT-D3. J Phys Chem B 118:3431–3440. https://doi.org/10.1021/jp411616b
Grimme S, Antony J, Ehrlich S, Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. https://doi.org/10.1063/1.3382344
Grimme S, Ehrlich S, Goerigk L (2011) Effect of the damping function in dispersion corrected density functional theory. J Comput Chem 32:1456–1465. https://doi.org/10.1002/jcc.21759
Becke AD, Johnson ER (2005) A density-functional model of the dispersion interaction. J Chem Phys. https://doi.org/10.1063/1.2065267
Sure R, Grimme S (2013) Corrected small basis set Hartree-Fock method for large systems. J Comput Chem 34:1672–1685. https://doi.org/10.1002/jcc.23317
Mikulskis P, Cioloboc D, Andrejić M et al (2014) Free-energy perturbation and quantum mechanical study of SAMPL4 octa-acid host-guest binding energies. J Comput Aided Mol Des 28:375–400. https://doi.org/10.1007/s10822-014-9739-x
Caldararu O, Olsson MA, Riplinger C et al (2017) Binding free energies in the SAMPL5 octa-acid host–guest challenge calculated with DFT-D3 and CCSD(T). J Comput Aided Mol Des 31:87–106. https://doi.org/10.1007/s10822-016-9957-5
Riplinger C, Pinski P, Becker U et al (2016) Sparse maps—a systematic infrastructure for reduced-scaling electronic structure methods. II. Linear scaling domain based pair natural orbital coupled cluster theory. J Chem Phys 144:024109. https://doi.org/10.1063/1.4939030
Grimme S (2006a) Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J Comput Chem 27:1787–1799. https://doi.org/10.1002/jcc.20495
Perdew JP, Wang Y (1992) Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B 45:13244–13249. https://doi.org/10.1103/PhysRevB.45.13244
Perdew JP, Chevary JA, Vosko SH et al (1992) Atoms, molecules, solids, and surfaces: applications of the generalized gradient approximation for exchange and correlation. Phys Rev B 46:6671–6687. https://doi.org/10.1103/PhysRevB.46.6671
Marenich AV, Cramer CJ, Truhlar DG (2009) Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J Phys Chem B 113:6378–6396. https://doi.org/10.1021/jp810292n
Grimme S (2006b) Semiempirical hybrid density functional with perturbative second-order correlation. J Chem Phys. https://doi.org/10.1063/1.2148954
Goerigk L, Grimme S (2011) Efficient and accurate double-hybrid-meta-GGA density functionals-evaluation with the extended GMTKN30 database for general main group thermochemistry, kinetics, and noncovalent interactions. J Chem Theory Comput 7:291–309. https://doi.org/10.1021/ct100466k
Mardirossian N, Head-Gordon M (2016) ωB97M-V: a combinatorially optimized, range-separated hybrid, meta-GGA density functional with VV10 nonlocal correlation. J Chem Phys. https://doi.org/10.1063/1.4952647
Mardirossian N, Head-Gordon M (2017) Thirty years of density functional theory in computational chemistry: an overview and extensive assessment of 200 density functionals. Mol Phys 115:2315–2372
Labute P (2009) Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins Struct Funct Bioinform 75:187–205. https://doi.org/10.1002/prot.22234
Chemical Computing Group Inc (2019) Molecular operating environment (MOE). Chemical Computing Group Inc., Montreal
Anthony WJ, Bender A, Kaya T, Clemons PA (2009) Alpha shapes applied to molecular shape characterization exhibit novel properties compared to established shape descriptors. J Chem Inf Model 49:2231–2241. https://doi.org/10.1021/ci900190z
Hoffmann R (1963) An extended Hückel theory. I Hydrocarbons. J Chem Phys 39:1397–1412. https://doi.org/10.1063/1.1734456
Hornak V, Abel R, Okur A et al (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725. https://doi.org/10.1002/prot.21123
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Corbeil CR, Williams CI, Labute P (2012) Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des 26:775–786. https://doi.org/10.1007/s10822-012-9570-1
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641. https://doi.org/10.1002/jcc.10128
Vanquelef E, Simon S, Marquant G et al (2011) R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res 39:511–517. https://doi.org/10.1093/nar/gkr288
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280. https://doi.org/10.1021/j100142a004
Case DA, Betz RM, Botello-Smith W et al (2018) Amber 18
Joung IS, Cheatham TE (2008) Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B 112:9020–9041. https://doi.org/10.1021/jp8001614
Horn HW, Swope WC, Pitera JW et al (2004) Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys 120:9665–9678. https://doi.org/10.1063/1.1683075
Döpke MF, Moultos OA, Hartkamp R (2020) On the transferability of ion parameters to the TIP4P/2005 water model using molecular dynamics simulations. J Chem Phys. https://doi.org/10.1063/1.5124448
Loncharich RJ, Brooks BR, Pastor RW (1992) Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 32:523–535. https://doi.org/10.1002/bip.360320508
Berendsen HJC, Postma JPM, Van Gunsteren WF et al (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Cerutti DS, Case DA (2010) Multi-level Ewald: a hybrid multigrid/fast fourier transform approach to the electrostatic particle-mesh problem. J Chem Theory Comput 6:443–458. https://doi.org/10.1021/ct900522g
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins Struct Funct Bioinform 55:383–394. https://doi.org/10.1002/prot.20033
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82. https://doi.org/10.1021/ci100275a
Frisch MJ, Trucks GW, Schlegel HB et al (2016) Gaussian 16 revision A03. Gaussian, Inc, Wallingford
Dunning TH (1989) Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J Chem Phys 90:1007–1023. https://doi.org/10.1063/1.456153
Smith DGA, Burns LA, Patkowski K, Sherrill CD (2016) Revised damping parameters for the D3 dispersion correction to density functional theory. J Phys Chem Lett 7:2197–2203. https://doi.org/10.1021/acs.jpclett.6b00780
Kendall RA, Dunning TH, Harrison RJ (1992) Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J Chem Phys 96:6796–6806. https://doi.org/10.1063/1.462569
Woon DE, Dunning TH (1993) Gaussian basis sets for use in correlated molecular calculations. III. The atoms aluminum through argon. J Chem Phys 98:1358–1371. https://doi.org/10.1063/1.464303
Dunning J, Peterson KA, Wilson AK (2001) Gaussian basis sets for use in correlated molecular calculations. X. The atoms aluminum through argon revisited. J Chem Phys 114:9244–9253. https://doi.org/10.1063/1.1367373
Hariharan PC, Pople JA (1973) The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chim Acta 28:213–222. https://doi.org/10.1007/BF00533485
Hehre WJ, Ditchfield K, Pople JA (1972) Self-consistent molecular orbital methods. XII. Further extensions of gaussian-type basis sets for use in molecular orbital studies of organic molecules. J Chem Phys 56:2257–2261. https://doi.org/10.1063/1.1677527
Tomasi J, Mennucci B, Cammi R (2005) Quantum mechanical continuum solvation models. Chem Rev 105:2999–3093. https://doi.org/10.1021/cr9904009
Merrick JP, Moran D, Radom L (2007) An evaluation of harmonic vibrational frequency scale factors. J Phys Chem A 111:11683–11700. https://doi.org/10.1021/jp073974n
Jensen JH (2015) Predicting accurate absolute binding energies in aqueous solution: thermodynamic considerations for electronic structure methods. Phys Chem Chem Phys 17:12441–12451. https://doi.org/10.1039/c5cp00628g
Klamt A, Moya C, Palomar J (2015) A comprehensive comparison of the IEFPCM and SS(V)PE continuum solvation methods with the COSMO approach. J Chem Theory Comput 11:4220–4225. https://doi.org/10.1021/acs.jctc.5b00601
Riplinger C, Neese F (2013) An efficient and near linear scaling pair natural orbital based local coupled cluster method. J Chem Phys. https://doi.org/10.1063/1.4773581
Neese F, Valeev EF (2011) Revisiting the atomic natural orbital approach for basis sets: robust systematic basis sets for explicitly correlated and conventional correlated ab initio methods? J Chem Theory Comput 7:33–43. https://doi.org/10.1021/ct100396y
Jensen F (2002) Polarization consistent basis sets: II. Estimating the Kohn-Sham basis set limit. J Chem Phys 116:7372–7379. https://doi.org/10.1063/1.1465405
Stück D, Baker TA, Zimmerman P et al (2011) On the nature of electron correlation in C60. J Chem Phys. https://doi.org/10.1063/1.3661158
Bauernschmitt R, Ahlrichs R (1996) Stability analysis for solutions of the closed shell Kohn-Sham equation. J Chem Phys 104:9047–9052. https://doi.org/10.1063/1.471637
Roos BO, Taylor PR, Sigbahn PEM (1980) A complete active space SCF method (CASSCF) using a density matrix formulated super-CI approach. Chem Phys 48:157–173. https://doi.org/10.1016/0301-0104(80)80045-0
Lee TJ, Taylor PR (1989) A diagnostic for determining the quality of single-reference electron correlation methods. Int J Quantum Chem 36:199–207. https://doi.org/10.1002/qua.560360824
Lee TJ, Rice JE, Scuseria GE, Schaefer HF (1989) Theoretical investigations of molecules composed only of fluorine, oxygen and nitrogen: determination of the equilibrium structures of FOOF, (NO)2 and FNNF and the transition state structure for FNNF cis-trans isomerization. Theor Chim Acta 75:81–98. https://doi.org/10.1007/BF00527711
Lee MC, Yang R, Duan Y (2005) Comparison between generalized-Born and Poisson-Boltzmann methods in physics-based scoring functions for protein structure prediction. J Mol Model 12:101–110. https://doi.org/10.1007/s00894-005-0013-y
Sure R, Grimme S (2015) Comprehensive benchmark of association (free) energies of realistic host–guest complexes. J Chem Theory Comput 11:3785–3801. https://doi.org/10.1021/acs.jctc.5b00296
Weigend F, Ahlrichs R (2005) Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys Chem Chem Phys 7:3297–3305. https://doi.org/10.1039/b508541a
Lee J, Head-Gordon M (2019) Distinguishing artificial and essential symmetry breaking in a single determinant: approach and application to the C60, C36, and C20 fullerenes. Phys Chem Chem Phys 21:4763–4778. https://doi.org/10.1039/c8cp07613h
Sherrill CD, Lee MS, Head-Gordon M (1999) On the performance of density functional theory for symmetry-breaking problems. Chem Phys Lett 302:425–430. https://doi.org/10.1016/S0009-2614(99)00206-7
Sherrill CD, Krylov AI, Byrd EFC, Head-Gordon M (1998) Energies and analytic gradients for a coupled-cluster doubles model using variational Brueckner orbitals: application to symmetry breaking in O4+. J Chem Phys 109:4171–4181. https://doi.org/10.1063/1.477023
Lee J, Head-Gordon M (2018) Regularized orbital-optimized second-order Møller-Plesset perturbation theory: a reliable fifth-order-scaling electron correlation model with orbital energy dependent regularizers. J Chem Theory Comput 14:5203–5219. https://doi.org/10.1021/acs.jctc.8b00731
Grimme S (2012) Supramolecular binding thermodynamics by dispersion-corrected density functional theory. Chemistry 18:9955–9964. https://doi.org/10.1002/chem.201200497
Li YP, Gomes J, Sharada SM et al (2015) Improved force-field parameters for QM/MM simulations of the energies of adsorption for molecules in zeolites and a free rotor correction to the rigid rotor harmonic oscillator model for adsorption enthalpies. J Phys Chem C 119:1840–1850. https://doi.org/10.1021/jp509921r
Acknowledgements
We gratefully acknowledge support from the University of North Texas Academic Computing Services for the use of the UNT Research Clusters and Institute for Cyber-Enabled Research (iCER) at Michigan State University. Computational resources were provided via the NSF Major Research Instrumentation program supported by the National Science Foundation under Grant No. CHE-1531468.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eken, Y., Almeida, N.M.S., Wang, C. et al. SAMPL7: Host–guest binding prediction by molecular dynamics and quantum mechanics. J Comput Aided Mol Des 35, 63–77 (2021). https://doi.org/10.1007/s10822-020-00357-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00357-3