Abstract
Falcipain-2 (FP-2) is a papain-family cysteine protease of Plasmodium falciparum whose primary function is to degrade the host red cell hemoglobin, within the food vacuole, in order to provide free amino acids for parasite protein synthesis. Additionally it promotes host cell rupture by cleaving the skeletal proteins of the erythrocyte membrane. Therefore, the inhibition of FP-2 represents a promising target in the search of novel anti-malarial drugs. A potent FP-2 inhibitor, characterized by the presence in its structure of the 1,4-benzodiazepine scaffold and an α,β-unsaturated methyl ester moiety capable to react with the Cys42 thiol group located in the active site of FP-2, has been recently reported in literature. In order to study in depth the inhibition mechanism triggered by this interesting compound, we carried out, through ONIOM hybrid calculations, a computational investigation of the processes occurring when the inhibitor targets the enzyme and eventually leads to an irreversible covalent Michael adduct. Each step of the reaction mechanism has been accurately characterized and a detailed description of each possible intermediate and transition state along the pathway has been reported.
Similar content being viewed by others
Introduction
Malaria is one of the most deadly parasitic diseases with an estimated 216 million clinical cases and nearly 655,000 deaths in 2010 [1]. Currently, 91 % of deaths happen in the southern of Africa, especially in children under the age of 5 years.
Malaria burden is caused by the Plasmodium parasite, transmitted through the bite of the infected female anopheline mosquitoes. The species able to infect humans are four: P. falciparum, P. vivax, P. malariae, and P. ovale. Among these, P. falciparum turns out to be the most lethal one and, together with P. vivax, accounts for more than 95 % of malaria cases in the world [2]. Moreover, in South East Asia, some infections due to P. knowlesi, a malaria parasite of monkey, have also been reported in humans [3].
In the last years, it has become evident that all classes of antimalarial drugs show resistance, such as the recently reported artemisinin [4, 5], limiting the efficacy of the applied therapy. Moreover, despite intense research efforts over the last two decades by big pharmaceutical industries and non profit organizations to develop a vaccine against malaria, to date it is not yet available. On the basis of these limitations, new targets for the design and development of antimalarial drugs are urgently needed [6].
A promising strategy to develop new drugs for the treatment of malaria is to target proteases of malaria parasites which are involved in the processes of host erythrocyte rupture, erythrocyte invasion, and hemoglobin degradation.
Attention has been particularly focused on falcipain-2 (FP-2), a papain-family cysteine protease that, within P. falciparum food vacuole, plays a primary role in the host hemoglobin digestion [7], that is essential for sustaining the metabolic needs of the parasite. Additionally, FP-2 is fundamental in the host membrane rupture required for the intracellular parasite release, being associated with the cleavage of erythrocyte membrane skeletal proteins, such as ankyrin and band 4.1 protein [8]. Hence, FP-2 is considered a promising target for the discovery of novel antimalarial drugs [9].
Many FP-2 inhibitors characterized by different structural features, i.e. peptides, peptidomimetics, isoquinolines, thiosemicarbazones, and chalcones, have been reported and proved to be capable to inactivate the enzyme in a reversible or irreversible manner [9].
In particular, some compounds, presenting in their structure a peptidyl moiety connected to an electrophilic warhead able to covalently bind the enzyme, showed low nM affinity and an irreversible inhibition of the enzyme. Among them, vinyl sulfones and aziridines [10] displayed a high potency, measured through the second-order rate constant of inhibition (k 2nd ) that is in the range 105–106 M−1 min−1. Recently, we synthesized a new series of peptidomimetics, based on the 1,4-benzodiazepine (1,4-BDZ) scaffold connected to different warheads, that demonstrated an even better biological activity [11]. Particularly interesting is the vinyl ester 1 that showed high affinity and potency (K i = 0.017 ± 0.004 μM, k 2nd = 3.57 ± 0.40 × 106 M−1 min−1) and resulted two times more potent than the reference compound carboxyoxiran E-64 (K i = 0.29 ± 0.09 μM, k 2nd = 1.70 ± 0.40 × 106 M−1 min−1) [7]. Compound 1 derived from several studies conducted modifying the general structure of typical FP-2 inhibitors, such as the vinyl sulfones, E-64, and the peptidyl aziridines inhibitors. Thus, 1 joins together a lipophilic aryl moiety able to increase the interaction with the enzyme P3 site, a central 1,4-BDZ moiety that limits the conformational freedom of the molecule, and a vinyl warhead able to covalently inhibit the enzyme (Scheme 1). Our structure–activity relationship (SAR) studies clarified that compound 1 represents the optimal structure able to block the enzyme [11–13]. Therefore, 1 has been identified as a new lead compound for the possible development of new antimalarial drugs.
A number of publications aiming to the prediction of the inhibition mechanism of cysteine–proteases, and structurally related enzymes, by means of computational approaches, have been published [14–18]. However, to the best of our knowledge, no theoretical studies have been conducted on FP-2. Thus, we decided to explore the mechanism of inhibition of compound 1 starting from the recently deposited crystal structure of FP-2 co-crystallized with E-64 [7] and taking into account that the α,β-unsaturated compounds act as irreversible inhibitors by means of a Michael reaction.
First, docking and molecular dynamics (MD) simulations were performed to obtain the protein/ligand complex. Then, a simplified molecular model, extracted from this system, was submitted to ONIOM hybrid calculation to investigate all the steps that characterize the inhibition mechanism operated by 1 by locating each predictable reaction intermediate and transition state, described in detail together with the related atomistic description.
Results and discussion
Systems setup by docking and MD simulations
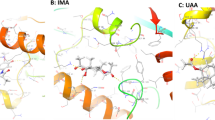
From the X-ray structure of FP-2 co-crystallized with the covalent inhibitor E-64 [7], a model of the enzyme was developed. The binding mode of E-64 was optimized by MD simulations. We noted that the cation-π interaction between the guanidine group of E-64 and the phenol ring of Tyr78 was displaced by a H-bond with the backbone of Asn82. Moreover, the H-bond between the carboxylic group of E-64 and Gln36 was replaced by three H-bonds with Cys42, Ser41 and His174 (Fig. 1a).
Binding modes of E-64 (A) and 1 (B) within the FP-2 catalytic site after MD simulations. Carbon atoms of the ligands are represented as stick and colored in green and cyan, respectively. The catalytic site of FP-2 is shown as gray surface and the hydrogen bonds are shown as orange dashed lines. The alignment of the electrophile within the known catalytic triad is also reported (C)
Then, using this optimized model of FP-2, deprived of E-64, docking calculations were performed to estimate the binding mode of compound 1. The results put in evidence that the inhibitor fits completely the catalytic crevice of the enzyme and creates a H-bond network with various FP-2 residues (Fig. 1b) [19].
In particular, the backbone of Gly83 and Ile85 behaved as a H-bond donor towards two carbonyl groups of 1, while the side chain of Asp234 acted as a H-bond acceptor with a NH group. Moreover, several hydrophobic contacts further stabilized the FP-2/ligand complex, i.e. the accommodation of the aryl moiety in a cleft shaped by the side chains of Phe236, Ile85, Leu84, and Asn86.
The electron withdrawing group (EWG) attached to the vinyl moiety, created a H-bond with the side chain of Trp206. In our hypothesis, this residue, conserved also in the sequence of FP-3, creates a crucial contact for the correct orientation of the ligand into the catalytic site of FP-2.
The complex resulting from docking calculations was optimized through molecular mechanics methods implemented in the Amber9 package [20] and, after a preliminary equilibration phase in which the conformation of the solvated complex was relaxed at 300 K, the geometrical stability was attained by 10 ns of MD simulations (Fig. 2). After this step, the binding mode of 1 resulted unchanged.
Therefore, to the aim of this work, a simplified system, obtained from the minimized structure extracted from the MD last frame, was submitted to ONIOM calculations, described in the next sections.
Mechanistic considerations
The enzyme inhibition mechanism operated by organic molecules can be theoretically described by hybrid methods. They have the advantage to take into consideration the effects of the solvent and the residues surrounding the catalytic site on the computation of the reaction mechanism.
Hybrid methods, such as the quantum mechanical/molecular mechanics (QM/MM) [21–29], are robust and accurate procedures but they are computationally expensive. On the other hand, valuable insights are also achievable from small and simplified models in which the side chains (or some atoms) of the catalytic triad of the enzymes are represented. For example, Paasche et al. [14] and Mladenovic et al. [18] studying thiol-containing enzymes, investigated the influence of the chemical structure of the inhibitors on the kinetic parameters of papain-like proteases adopting model systems containing a simplified structure of the inhibitors, ammonia and methanethiol [14, 18, 30].
Recently, to rationalize the computer time requested by QM/MM approaches and to increase the details of the simulation systems, the ONIOM hybrid approach [31–33] (Gaussian09 package) [34] has been applied to theoretically study enzyme catalysis [15]. The calculations have been performed on simplified models of the complexes including the inhibitors warhead together with the amino acids shaping the catalytic site.
Hence, we built a model system (Fig. 3) including the catalytic residues [35] Cys42, His174, Asn204, Trp206 together with the atoms contained in a sphere of 4Å centered on the ligand vinyl warhead. The structure of the ligand was truncated in proximity of the peptide bond internal to the 1,4-BDZ ring.
Representation of the model system employed for the computation of the inhibition mechanism operated by the vinyl-ester 1. The high layer of ONIOM is represented as stick model while the low layer is represented as lines. The represented snapshot shows the complex after the optimization step at B3LYP/6-31G(d)/AM1 level of theory
ONIOM calculations offered the possibility to consider as High Layer (HL) the atoms partecipating to the reaction while the remaining atoms were considered as Low Layer (LL). Density functional theory (DFT) at the B3LYP/6-31G(d) level [36, 37] was used in the energy optimization step for the HL while the LL was calculated at the AM1 level [38]. Then, to obtain more accurate energy values, the system was submitted to single-point calculations in which the HL was treated at the M06/6-31++G(3df,2pd) [39] level and the solvent effects were estimated through IEFPCM [40] calculations in water.
Starting from the geometry obtained by the docking and MD approaches, the conformations of the various intermediates and their related TSs were manually built. The system was simulated without the interference of proton or solvent molecules. This is in agreement with the experimental FP-2 kinetic data showing that FP-2 was in a “cleaving” active state in a pH range from 4.5 to 8.5, with an optimal value of 7.5 [35]. Additionally, the results obtained for the thiol-containing SARS-CoV protease, an enzyme belonging to the FP-2 family, are in line with our proposals [14]. Furthermore, looking into our complex, the catalytic amino acidic triad (Cys42, His174, Asn204) appeared protected from the contact of the solvent by the presence of the bulky structure of the 1,4-BDZ substrate.
Several possible intermediates could be envisaged and located; in the low energy reaction pathway Cys42, deprotonated by His174, acted as a nucleophile capable to attack Cβ of the α,β-unsaturated compound to yield an enolate, as occurs in the classical Michael reaction. In our hypotheses, a rotation around the Cα–Cβ single bond allowed the enolate to assume a new conformation, able to be protonated at Cα thus giving the final Michael adduct (Scheme 2).
Computations
The covalent inhibition started when the inhibitor entered the active site of the enzyme to form the complex FP-2/1 in a geometry (S) in which the amino acids of the catalytic triad were perfectly aligned with His174 acting both as a H-bond donor and an acceptor toward Asn204 and Cys42, respectively. In addition, the side chain of Trp206 created a H-bond with the carbonyl group belonging to the ester function of 1 (Scheme 2; Fig. 4), increasing the electrophilicity of the ligand, as shown in Fig. 5.
Reaction pathway calculated at the M06/6-31++G(3df,2pd): AM1-IEFPCM level for the covalent inhibition of FP-2 by the vinyl-ester (1). The relative energies are expressed as kcal/mol, the carbon atoms of 1 are colored in green while those of the enzyme are gray. Only the atoms involved in the reaction are shown for sake of clarity. The black continuous line highlights route B of Scheme 2, while the cyan dotted line shows route A
Then, His174, working as a base, deprotonated the thiol group of Cys42 yielding the thiolate (Cys42−) and the histidinium (His174+) ions (intermediate T). His174+ was still joined to the carbonyl group of Asn204 by a H-bond (1.75 Å) and the carbonyl group of the ligand to the side chain of Trp206 (2.10 Å). Moreover, the H-bond between Cys42− and His174+ is still maintained with reversed donor/acceptor roles (2.02 Å) with the sulfur atom of Cys42− also accepting a H-bond from the backbone NH of Ala175 (2.63 Å); conversely, this sulfur atom is far from Cβ of 1 (4.46 Å).
This step did not lead to a remarkable change in the relative energy of the system (ΔE = 2.6 kcal/mol) and, from a mechanistic point of view, T could be obtained from S by a hydrogen transfer taking place over an energy barrier of 5.3 kcal/mol (TS1). In the saddle point, the movable hydrogen was closer to the basic nitrogen belonging to the imidazole ring of His174 (1.28 Å) than the sulfur atom of Cys42 (1.69 Å).
From T, the sulfur anion of Cys42− could react with Cβ of the α,β-unsaturated ester (1), via an energy barrier of 5.7 kcal/mol (TS2), to yield the enolate EA. In this energetic costly step, Cys42− worked as a soft nucleophile capable of attacking the positively polarized β-carbon of 1. During the sulfur attack, a concerted movement of His174+, that rotated one of its NH away from the sulfur atom towards the ligand carbonyl oxygen with formation of a new H-bond, favored a further impoverishment of the electron density at Cβ.
Then, EA could be reversibly converted into enol E (route A on Scheme 2) or, alternatively, could assume, through a rotation around the Cα–Cβ single bond, a new conformation, EB, characterized by a different H-bond network (route B on Scheme 2). Actually, whereas EA maintained the almost planar arrangement characteristic of the parent α,β-unsaturated system (τCγ–Cβ–Cα–C(O) = 174°), in EB the same torsional angle τ assumed a value of −118°. Moreover, in EB His174+ had moved its NH away from the (former) carbonyl oxygen and established a new H-bond with the partially negative Cα atom of the ligand characterized by a distance of 1.81 Å (3.25 Å in EA). The other H-bonds were maintained, i.e. that of the carbonyl oxygen with the side chain of Trp206 and that of His174+ with Asn204. Both the higher stability of EB (ΔE = −1.2 kcal/mol) and the low energy barrier of 1.5 kcal/mol (TS4) between EA and EB ensured their easy interconversion. Conversely, the energy of E was higher than EA (ΔE = 1.5 kcal/mol) and the energy barrier was 2.0 kcal/mol (TS3). It is worthy to note that only EB could be converted into the final adduct K whereas E had to go back to the enolates prior to be converted into its keto-tautomeric form, actually corresponding to the final adduct K. So, route A was not only energetically disfavored but it led to a structure (E) not directly linked to the final adduct.
In the final step EB was converted into the final product K over the very low energy barrier TS5 (0.2 kcal/mol). The transition state, governing the chemical switch, required a shift of the hydrogen atom from the imidazole ring of His174+ to the Cα atom of 1. The relative energy of TS5, very close to that of EB, made this shift straightaway when EB was obtained from EA. The overall process was exoergonic as the final Michael adduct K resulted 17.3 kcal/mol lower in energy than the starting complex S.
Experimental section
Docking and MD
The structure of FP-2, retrieved from the Protein Data Bank (code 3BPF), was geometrically refined following the molecular modeling standard procedures: minimization, equilibration and molecular dynamics simulation, previously optimized by us [41, 42]. Ligands were built by Sybyl 8.0 [43] and preliminarily minimized by Gaussian09 [34] at the DFT/B3LYP/6-31G(d) level. Docking calculations were then performed with the GOLD 5.0 program [44] into the binding cleft depicted by the presence of E-64. The cavity was detected with an active site radius of 20.0 Å from the side chain sulfur atom of residue Cys42. The side chain of Trp206 was imposed movable during the docking calculation. The goldscore fitness function and the distribution of torsional angles were chosen to evaluate the quality of the docking results. Van der Waals and hydrogen bonding radii were set at 4.0 and 3.0 Å, respectively; genetic algorithm parameters were kept at the default value. The complex formed by FP-2 and the highest scored docking solution was submitted to molecular dynamics (MD) simulations with the SANDER module of the AMBER9 package [20]. The ff03 and GAFF force fields were applied for the protein and the ligand, respectively. The complex was immersed in a box containing about 1,000 water molecules and the TIP3P model [45] was employed to explicitly represent the solvent. Na+ counterions were added to maintain neutrality of the system. At first, the energy of the water molecules was minimized, keeping the atoms of the enzyme frozen. Then, a minimization of the whole system was performed by setting a convergence criterion on the gradient of 10−4 kcal mol−1 Å−1. Prior to starting the MD simulations, the system was equilibrated for 40 ps at 300 K in isocore conditions (NVT). Subsequently, 10 ns of MD simulations in isothermal-isobaric ensemble were carried out at 300 K with a 2 fs time-step (NPT). In the production runs, the system was performed in periodic boundary conditions. Van der Waals and short-range electrostatic interactions were estimated within a 8 Å cutoff. SHAKE algorithm was applied to all bonds involving hydrogen atoms. When the geometrical stabilization of the complex was reached, a new minimization of the whole system was performed. On this snapshot was applied the subsequent ONIOM calculations.
ONIOM calculations
We extracted a simple model system from the FP-2/1 complex, including the triad catalytic residues Cys42, His174, Asn204 and Trp206 together with the atoms contained in a sphere of 4 Å centred on the ligand. No water molecules near the binding site, involved in H-bonding interactions with the ligand or the pocket, were observed. The stereochemistry of the covalent adduct had been chosen taking into account the structure–activity relationships reported in literature [11].
All the calculations were carried out using the Gaussian09 program package [34]. To rationalize the computational time, we have resorted to the ONIOM hybrid method [31–33] in the geometry optimization. Density functional theory (DFT) with the B3LYP [36, 37] functional and the 6-31G(d) basis set was used in all geometry optimizations. The low layer, kept fixed, was treated with the semiempirical method AM1 [38]. The high layer contained the atoms belonging to the ligand and the complete structures of Cys42, His174, Asn204, and Trp206, and the low layer enclosed the surrounding atoms of the catalytic site. Hydrogen atoms were used as link atoms at the truncated bonds. Vibrational frequencies were computed at the same level of theory to define the optimized structures as minima or transition states, which present an imaginary frequency corresponding to the forming bonds.
The stationary points’ final energies were recalculated through single point calculations at the M06/6-31++G(3df,2pd)/AM1 level [39]. The solvent effects were considered at the same level as above, on the gas-phase optimized geometries, using a self-consistent reaction field (SCRF) method, based on the Integral-Equation-Formalism Polarizable Continuum Model (IEFPCM) [40].
Figures were acquired by the PyMOL software (The PyMOL Molecular Graphics System, Version 0.99).
Conclusions
In this effort, by means of computational studies, we highlighted the FP-2 inhibition mechanism operated by the potent α,β-unsaturated 1,4-BDZ methyl ester 1. We applied a classical docking/MD protocol to build the ligand/enzyme complex and then the ONIOM module to acquire the atomic details and the energy values of the different intermediates leading to the final Michael adduct.
The reaction between the enzyme and the inhibitor starts with the deprotonation of the thiol group of Cys42 by the side chain of His174. Then, the thiolate, acting as a nucleophile, attacks Cβ of the α,β-unsaturated compound giving the enolate A (EA). A rotation around the Cα–Cβ single bond of the ligand, allows EA to assume a new conformation (enolate B, EB), able to be protonated at Cα giving the final Michael adduct (K). Compound 1 proceeds to the formation of K overcoming the rate-limiting energy barrier (TS2) and passing through several intermediates with decreasing energy values.
Our results highlight that, in addition to the known catalytic triad, Trp206 plays a fundamental role in the anchoring of the ligand warhead, as well as in triggering the inhibition process of the enzyme. In fact, Trp206 firstly increases the Cβ electrophilicity of the ligand assisting the sulphur attack and then stabilizes the enolate intermediates.
These results shed light on the inhibition mechanism operated by this interesting α,β-unsaturated system, supporting the design of novel compounds inhibiting the FP-2 enzyme, a molecular target of high impact in the medicinal chemistry research.
References
World Malaria Report (2011) http://www.who.int/malaria/world_malaria_report_2011/en/
Ashley E, McGready R, Proux S, Nosten F (2006) Malaria. Travel Med Infect Dis 4:159–173
Singh B, Lee KS, Matusop A, Radhakrishnan A, Shamsul SSG, Cox-Singh J, Thomas A, Conway DJ (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363:1017–1024
Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ (2009) Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467
Dondorp AM, Yeung S, White L, Nguon C, Day NPS, Socheat D, von Seidlein L (2010) Artemisinin resistance: current status and scenarios for containment. Nat Rev Microbiol 8:272–280
Jana S, Paliwal J (2007) Novel molecular targets for antimalarial chemotherapy. Int J Antimicrob Ag 30:4–10
Kerr ID, Lee JH, Pandey KC, Harrison A, Sajid M, Rosenthal PJ, Brinen LS (2009) Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity. J Med Chem 52:852–857
Hanspal M, Dua M, Takakuwa Y, Chishti AH, Mizuno A (2002) Plasmodium falciparum cysteine protease falcipain-2 cleaves erythrocyte membrane skeletal proteins at late stages of parasite development. Blood 100:1048–1054
Ettari R, Bova F, Zappalà M, Grasso S, Micale N (2010) Falcipain-2 inhibitors. Med Res Rev 1:136–167
Schulz F, Gelhaus C, Degel B, Vicik R, Heppner S, Breuning A, Leippe M, Gut J, Rosenthal PJ, Schirmeister T (2007) Screening of protease inhibitors as antiplasmodial agents. Part I: aziridines and epoxides. ChemMedChem 2:1214–1224
Ettari R, Micale N, Schirmeister T, Gelhaus C, Leippe M, Nizi E, Di Francesco ME, Grasso S, Zappalà M (2009) Novel peptidomimetics containing a vinyl ester moiety as highly potent and selective falcipain-2 inhibitors. J Med Chem 52:2157–2160
Ettari R, Nizi E, Di Francesco ME, Dude M, Pradel G, Vicik R, Schirmeister T, Micale N, Grasso S, Zappalà M (2008) Development of peptidomimetics with a vinyl sulfone warhead as irreversibile falciai-2 inhibitors. J Med Chem 51:988–996
Ettari R, Nizi E, Di Francesco ME, Micale N, Grasso S, Zappalà M, Vičík R, Schirmeister T (2008) Nonpeptidic vinyl and allyl phosphonates as falcipain-2 inhibitors. ChemMedChem 3:1030–1033
Paasche A, Schiller M, Schirmeister T, Engels B (2010) Mechanistic study of the reaction of thiol-containing enzymes with α, β-unsaturated carbonyl substrates by computation and chemoassays. ChemMedChem 5:869–880
Dourado DFAR, Fernandes PA, Ramos MJ (2010) Glutathione transferase classes alpha, pi, and mu: GSH activation mechanism. J Phys Chem B 114:12972–12980
Fernandes PA, Ramos MJ (2003) Theoretical studies on the mechanism of inhibition of ribonucleotide reductase by (E)-2′-fluoromethylene-2′-deoxycitidine-5′-diphosphate. J Am Chem Soc 125:6311–6322
Schwöbel JAH, Wondrousch D, Koleva YK, Madden JC, Cronin MTD, Schüürmann G (2010) Prediction of Michael-type acceptor reactivity toward glutathione. Chem Res Toxicol 23:1576–1585
Mladenovic M, Schirmeister T, Thiel S, Thiel W, Engels B (2007) The importance of the active site histidine for the activity of epoxide- or aziridine-based inhibitors of cysteine proteases. ChemMedChem 2:120–128
Ettari R, Zappalà M, Micale N, Grazioso G, Giofrè S, Schirmeister T, Grasso S (2011) Peptidomimetics containing a vinyl ketone warhead as falcipain-2 inhibitors. Eur J Med Chem 46:2058–2065
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9. University of California, San Francisco
Aaqvist J, Warshel A (1993) Simulation of enzyme reactions using valence bond force fields and other hybrid quantum/classical approaches. Chem Rev 93:2523–2544
Gao J (1995) In: Lipkowitz KB, Boyd DB (eds) Reviews in computational chemistry. VCH Publishers, New York
Mordasini TZ, Thiel W (1998) Computational chemistry column: combined quantum mechanical and molecular mechanical approaches. Chimia 52:288–291
Monard G, Merz KM Jr (1999) Combined quantum mechanical/molecular mechanical methodologies applied to biomolecular systems. Acc Chem Res 32:904–911
Gao J, Truhlar DG (2002) Quantum mechanical methods for enzyme kinetics. Annu Rev Phys Chem 53:467–505
Field MJ (2002) Computational studies of reaction mechanisms of methane monooxygenase and ribonucleotide reductase. J Comp Chem 23:48–58
Monard G, Prat-Resina X, González-Lafont A, Lluch JM (2003) Determination of enzymatic reaction pathways using QM/MM methods. Int J Quantum Chem 93:229–244
Ridder L, Mulholland A (2003) Modeling biotransformation reactions by combined quantum mechanical/molecular mechanical approaches: from structure to activity. Curr Top Med Chem 3:1241–1256
Sgrignani J, Bonaccini C, Grazioso G, Chioccioli M, Cavalli A, Gratteri P (2009) Insights into docking and scoring neuronal α4β2 nicotinic receptor agonists using Molecular Dynamics simulations and QM/MM calculations. J Comp Chem 30:2443–2454
Paasche A, Arnone M, Fink R, Schirmeister T, Engels B (2009) Origin of the reactivity differences of substituted aziridines: CN vs CC bond breakages. J Org Chem 74:5244–5249
Bakowies D, Thiel W (1996) Hybrid models for combined quantum mechanical and molecular mechanical approaches. J Phys Chem 100:10580–10594
Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ (1999) A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J Mol Struct Theochem 462:1–21
Cosman PC, Rogers JK, Sherwood PG, Zeger K (2000) Combined forward error control and packetized zerotree wavelet encoding for transmission of images over varying channels. IEEE Trans Image Process 9:982–993
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery Jr JA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, Revision A.02. Gaussian, Inc., Wallingford
Hogg T, Nagarajam K, Herzberg S, Chen L, Shen X, Jiang H, Wecke M, Blohmke C, Hilgenfeld R, Schmidt C (2006) Structural and functional characterization of falcipain-2, a hemoglobinase from the malarial parasite Plasmodium falciparum. J Biol Chem 281:25425–25437
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785–789
Dewar MJS, Zoebisch EG, Healy EF (1985) AM1: a new general purpose quantum mechanical molecular model. J Am Chem Soc 107:3902–3909
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functional. Theor Chem Acc 120:215–241
Cancés E, Mennucci B, Tomasi J (1997) A new integral equation formalism for the polarizable continuum model: theoretical background and applications to isotropic and anisotropic dielectrics. J Chem Phys 107:3032–3042
Grazioso G, Cavalli A, De Amici M, Recanatini M, De Micheli C (2008) Alpha7 nicotinic acetylcholine receptor agonists: prediction of their binding affinity through a molecular mechanics poisson-boltzmann surface area approach. J Comp Chem 29:2593–2602
Grazioso G, Pomè DY, Matera C, Frigerio F, Pucci L, Gotti C, Dallanoce C, De Amici M (2009) Design of novel alpha7-subtype-preferring nicotinic acetylcholine receptor agonists: application of docking and MM-PBSA computational approaches, synthetic and pharmacological studies. Bioorg Med Chem Lett 19:6353–6357
Sybyl 8.0, Tripos International, 1699 South Hanley Rd., St. Louis, Missouri 63144, USA
Verdonk ML, Cole JC, Hartshorn M, Murray CW, Taylor R (2003) Improved protein-ligand docking using GOLD. Proteins 52:609–623
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein LM (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–936
Acknowledgments
This work was supported by the Italian “Ministero dell’Istruzione, dell’Università e della Ricerca” and by the Universities of Pavia (FAR grant) and Milan (PUR grant). LL and GG are very grateful to Professor Pierluigi Caramella for helpful discussions. CILEA (Milan) and CASPUR (Rome) are acknowledged for allocation of computer time. RE acknowledges “Dote Ricerca: FSE, Regione Lombardia” for supporting her postdoc fellowship. GG dedicate this effort to the memory of Dr. Agata Annino, who left us seven years ago.
Author information
Authors and Affiliations
Corresponding author
Additional information
Giovanni Grazioso and Laura Legnani contributed equally to the work.
Rights and permissions
About this article
Cite this article
Grazioso, G., Legnani, L., Toma, L. et al. Mechanism of falcipain-2 inhibition by α,β-unsaturated benzo[1,4]diazepin-2-one methyl ester. J Comput Aided Mol Des 26, 1035–1043 (2012). https://doi.org/10.1007/s10822-012-9596-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-012-9596-4