Abstract
Successful aquatic biological invasions are often dependent on human vectors. The Chinese pond mussel Sinanodonta woodiana is a fast-growing, high-filtering, and highly fecund unionid bivalve that has expanded rapidly throughout Europe in the last several decades. Human-mediated vectors such as fish aquaculture and pet shop trade are believed to be primary drivers of the mussel’s spread, but the specific dispersal pathways of S. woodiana remain unclear. We analyzed eleven microsatellites in 245 S. woodiana individuals from seven wild populations and three pet shops to assess genetic relatedness potential sources, and dispersal pathways of S. woodiana in Bavaria, Germany. Wild populations shared a single mitochondrial haplotype present in all European populations and shop-sourced individuals were genetically similar to wild populations. Only one wild population was structurally different than all other populations, which may be explained by its dispersal trajectory. Our results provide the first genetically informed insights into S. woodiana dispersal in the context of human activity. We underscore the value of genetic tools in invasive species management, which can uncover patterns of dispersal even on local scales.
Similar content being viewed by others
Introduction
Climate change and human activity have facilitated the spread of invasive species on a global scale. Aquatic systems are highly permeable, highly degraded by human activity, and are thus at high risk of invasion by mobile generalist species (Light and Moyle 2015). Due to their strong, multiple, and negative effects on native biodiversity, invasive species are considered one of the most significant threats to aquatic systems worldwide (Geist 2011; Geist et al. 2023).
Some of the most successful aquatic invaders are freshwater bivalves. In Europe and North America, the zebra mussel Dreissena polymorpha (Pallas 1771) and the Asian clam Corbicula fluminea (Mueller 1774) dominate community biomass in many freshwater systems (Strayer et al. 1996). Their success is attributed not only to their competitive biology but also their use of human vectors such as boating and shipping, by which the species primarily spread.
Successful aquatic biological invasions are often dependent on human vectors, where humans are often unaware of their facilitative role. As invasive “flagship” species, D. polymorpha and C. fluminea are easily recognizable, observed in high densities, and are thus common targets for management programs (Sousa et al. 2014). This is not the case for other, lesser-known species, whose invasion trajectories, while similarly linked to human activity, remain much less understood.
The Chinese pond mussel Sinanodonta woodiana (Lea 1834) is a fast-growing, high-filtering unionid bivalve which spread rapidly from its native East-Asian range into southern Asia, Europe, and the Americas in the 1960s (Konečný et al. 2018). In Europe, the mussel is found in fish ponds and heated lake systems as well as natural lakes, rivers, and slow-moving streams (Urbańska et al. 2021; Dobler et al. 2022; Geist et al. 2023), in a range of temperatures and substrate types (Spyra et al. 2016; Poznańska-Kakareko et al. 2021). First investigations of the species have shown that S. woodiana grows at fast rates (Dudgeon and Morton 1983; Sárkány-Kiss et al. 2000) and produces multiple larvae broods per year in high numbers (Wachtler et al. 2001; Labecka and Domagala 2018; Labecka and Czarnoleski 2021). The mussel also infests host fishes at higher rates compared to native mussels (Huber and Geist 2019), induces cross-resistance and elevates stress responses in host fishes (Donrovich et al. 2017; Reichard et al. 2022), competitively partitions food sources (Douda and Čadková 2018), outcompetes native mussels for space (Poznańska-Kakereko et al. 2021; Urbańska et al. 2021), and can tolerate higher levels of anthropogenic stress than native mussels (Bielen et al. 2016). Its morphology, which is similar to that of native European pond mussels such as the duck mussel Anodonta anatina (Linnaeus 1758) or the swan mussel Anodonta cygnea (Linnaeus 1758), also likely contributes to its invasion success, as it is often misidentified as native (Guarneri et al. 2014; Karaouzas et al. 2022) and left to spread in natural water bodies.
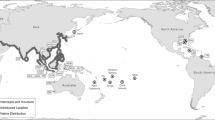
One of the most established aspects of the S. woodiana invasion is that the mussel spreads via fish hosts, and that this spread is largely facilitated by fish aquaculture (Urbańska et al. 2021). All European populations are believed to have been established from a primary import of larvae-infested fishes from East Asia, where the mussel then spread throughout a highly connected aquaculture network (Konečný et al. 2018). S. woodiana has been documented widely in carp ponds across Europe (Gabka et al. 2007; Najberek et al. 2011; Andrzejewski et al. 2013; Dobler et al. 2022), where their populations can spread rapidly via multiple dispersal pathways (Fig. 1). Carp and other fish species of commercial importance for aquaculture and recreational fishing may be transferred between ponds with S. woodiana larvae already encysted on their gills or fins (Soroka et al. 2014). Infested carp may also be maintained with other fishes used for stocking natural water bodies (Brämick 2019; Dobler et al. 2022). Infested fishes or free-living larvae, which remain viable without a fish host for weeks in a range of temperatures (Benedict and Geist 2021), may also be released into natural water bodies during annual pond draining events.
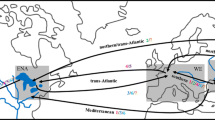
Fish aquaculture is often cited as the primary vector of the mussel’s spread (Bolotov et al. 2016; Kondakov et al. 2020; Urbańska et al. 2021), but this has not been systematically confirmed in comparison to other potential vectors. On the other hand, pet shop trade is also considered a major facilitator of biological invasions (Hulme et al. 2008; Nunes et al. 2015), but its role in the context of the S. woodiana invasion remains unclear. Only a few studies have mentioned pet shops as a potential vector of the mussel’s spread (Schoolmann et al. 2006; Bahr and Wiese 2018; Huber and Geist 2019; Dobler et al. 2022). These studies indicate that shops sell a variety of mussels within a singular category, often labeled as “European pond mussels,” although the category usually contains two to three species with similar habitat requirements and morphologies such as A. anatina, A. cygnea, and S. woodiana. Anecdotal evidence from shop websites shows that S. woodiana individuals are often sold in disguise of native mussels, sold in bulk, and directly released into ponds and backyard streams at high numbers (Fig. 2).
Selected images from popular pet shops that show mussels for sale. a Two S. woodiana specimens falsely advertised as native “XXL” or “normal” pond mussels. b A collection of S. woodiana advertised as pond mussels “sourced from Europe.” Images sourced from (a) Teichzeit, NatureHolic GmbH, https://www.teichzeit.de and (b) FördeFisch, Fa. Fördefisch, https://foerdefisch-online.de
Genetic tools are ideal for accurately identifying mussels to species level, including their early life stages (Zieritz et al. 2012). They can also be used to answer questions of dispersal and uncover how organisms move across space. Most genetic investigations of S. woodiana have occurred only in the last ten years and are generally limited to sequencing or RFLP analyses of the COI and ITS regions (Soroka 2005; Guarneri et al. 2014; Soroka et al. 2014; Bolotov et al. 2016). Sequencing of COI in S. woodiana has revealed a single haplotype (HapE-3) in all sampled European populations, and RFLP analyses of COI have confirmed low variation in parallel (Konečný et al. 2018; Soroka 2005). To our knowledge, only one study has used microsatellite genotyping to track the S. woodiana invasion across Europe, which found high levels of gene flow across the continent (Konečný et al. 2018).
These studies are useful for understanding the general demography and relatedness of European S. woodiana populations, but they are not representative of the entirety of Europe as few studies include western populations, including those in Germany. In addition, these studies are not sufficient for understanding the relatedness of populations on shorter time scales (i.e., local dispersal). There currently exists no study that uses genetic tools to investigate how S. woodiana disperses so rapidly outside of its native range, particularly within the context of human activity, which is believed to be the primary vector of the mussel’s spread. Because of this, many gaps remain in the knowledge of the mussel’s European demography, and many assumptions about its spread, especially on local scales and dispersal modes, remain untested.
The main objectives of this study were to 1) establish the first genetic records for S. woodiana in Germany and thus fill the knowledge gap for Europe-wide studies on the mussel, and to 2) use microsatellite genotyping to assess the genetic relationships between seven wild and three shop-sourced populations of S. woodiana in Bavaria, Germany.
Materials and methods
Mussel collection
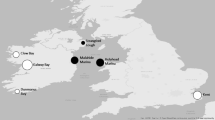
In June 2022, S. woodiana individuals (mean N = 24/shop) were ordered from the three most prominent pet shops in Germany using the search terms “pond mussels” or “pond mussels from Europe”. Shops were chosen based upon their popularity and visibility using internet search engines. All mussels were identified visually to species level and a foot tissue sample from each individual was collected, stored in RNAlater reagent, and frozen at − 20 °C until further analysis. Additional S. woodiana individuals were collected in the summers of 2020 and 2021 from seven known populations (n = 245) in Bavaria, Germany from a range of habitats in the Danube and Main/Rhine Basins (Fig. 3, Table 1). At each site, mussels were collected by randomly distributing 0.5 m × 0.5 m or 1 m × 1 m sample frames in near-shore soft substrates (max. 4 m depth) and retrieving all mussels within each frame. In sites with high turbidity or with a depth > 1 m, scuba and snorkeling methods were used to place frames and retrieve mussels. All mussels were identified visually to the species level and a foot tissue sample from each individual was collected, stored in RNAlater reagent, and frozen at − 20 °C until further analysis.
DNA extraction and genotyping
DNA was extracted from 25 mg of each tissue sample using the Macherey–Nagel NucleoSpin Tissue Kit following manufacturer protocol. Pure eluted DNA was diluted using a Nanodrop ND-100® spectrophotometer to establish working solutions containing 3–20 ng/µl of DNA product. To confirm the species identity of our wild populations, a 660–685 bp segment of the mitochondrial COI gene was amplified in a subset of samples (n = 8 from 4 populations) using the Qiagen Multiplex PCR kit (Qiagen®) in a PCR mix of 100 ng of cellular DNA, 10 pmol/µl each of LCO14 and HCO2198 primers (Konečný et al. 2018), 6 µl of Qiagen-Mix, and 5 µl of water for a final volume of 13 µl. All reactions were run using a Biometra UNO-11 cycler with an annealing temperature of 51 °C. PCR products were separated on 1.8% agarose gels via gel electrophoresis and extracted using the Machery-Nagel gel purification kit. Purified DNA from all samples was sequenced by Azenta Life Sciences using reverse primers. Raw sequence reads were cleaned and analyzed using Geneious Prime Version 10.2.6.
A set of 11 nuclear microsatellite markers identified by Popa et al. 2011 and 2015 (AW28, AW238, AW521, AW570, AW292, SW2, SW3, SW7, SW13, SW15, SW18) were amplified using the Qiagen Multiplex PCR kit (Qiagen®) in a PCR mix containing 100 ng of cellular DNA, 0.10–0.36 pmol/µl of primers, 7.5 µl of Qiagen-Mix, and 5.7–6.0 µl of water. Forward primers were labeled with TAMRA, HEX, and 6-FAM fluorescent dyes. PCR product was diluted to 0.5 µl and combined with a 1.7 µl mixture containing 56 µl of Formamide buffer and 12 µl of ROX standard, denatured at 94 °C for three minutes, cooled on ice, and loaded into an ABI Prism 310 Genetic Analyzer (Applied Biosystems, CA, USA) with a GENESCAN-500 (LIZ) internal size standard following methods from Geist and Kuehn (2005). A reference sample was included in each run to ensure exact scoring and cross-referencing between gels. GeneScan® and Genotyper® softwares (Version 2.5, Applied Biosystems, CA, USA) were used for allele scoring.
Statistical analyses
Geneious Prime (version 10.2.6; Biomatters Ltd., New Zealand) was used to determine pairwise percent identity between our COI reads and reference samples from NCBI. GENEPOP 4.0 (Rousset 2008) was used to calculate allele frequencies, average allele numbers per locus (A), expected and observed heterozygosities (HE, Ho), and pairwise FST values (Weir and Cockerham 1984). Loci with high allelic diversity can suppress FST values and lead to an overestimation of relatedness between groups. To account for this potential bias, Jost’s D (Jost 2008) values were calculated using GenAlEx 6.5 (Peakall and Smouse 2012). GENEPOP was also used to test for significant population differentiation among all pairs of populations using 100,000 iterations and 1000 de-memorisation steps (Raymond and Rousset 1995), and to test each locus in each population for conformance with Hardy–Weinberg (HW) expectations. To visualize levels of relatedness between populations, the kmeans function in the stats package in R (R Core Team, 2022) was used to group FST and Jost’s D values into three clusters (centers = 3) using 25 configurations. This visualization is represented as three shades which represent levels of relatedness (low, intermediate, and high) in Table 4. FSTAT 2.9.3 (Goudet 2001) was used to calculate values of FIS and allelic richness (AR), which was corrected by sample size. Private alleles were identified if they showed a frequency of more than 5% in one population and did not occur in any other population. 2MOD was used to estimate relatedness between individuals using the F value (Ciofi and Bruford 1999). MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004) was used to check the data for genotyping errors and null alleles. The impact of null alleles on genetic differentiation of populations was evaluated with the software FREENA (Chapuis and Estoup 2007) by comparing FST estimates before and after correcting for null alleles. To visualize the structure of our populations, we conducted a discriminant analysis of principal components (DAPC) using the dapc function in the ADEGENET v. 1.3–5 package in R (Pritchard et al. 2000; Jombart et al. 2010; R Core Team, 2022). This function identifies potential groups of relatedness by first transforming the data by principal component analysis before clustering by discriminant analysis. We retained 10 principal components and 3 discriminant functions. Similar colors correspond to similar genetic groupings of respective individuals and populations.
Results
Lineage assessment of wild populations
A multiple pairwise alignment of seven COI sequences from four wild populations to five reference genes produced a consistent 99.9% pairwise identity match to S. woodiana individuals sourced from Europe (Table 2). COI sequence alignments were 370 bp in length with no gaps and no polymorphisms. European populations belong to the HapE-3 lineage, and it is assumed that this lineage is represented in the selected reference genes.
Genetic diversity
MICROCHECKER did not detect any genotyping errors among the dataset but revealed signs of possible null alleles in all populations and at all loci except three (AW238, SW3, and AW521). These potential null alleles had no or negligible impact on genetic differentiation of populations as determined by FREENA. Because of this, the data set was not corrected for null alleles for subsequent analyses. Table 3 provides a summary of all indices of microsatellite diversity, including population codes.
The microsatellite markers applied in this study showed that pet shop populations exhibit an allelic richness (AR) of 4.3–4.7 (Table 3). The average number of alleles per locus (A) was similar between all three shop populations (A = 4.5, 5.0, and 5.0; AR = 4.3, 4.7, and 5.0 for FF, TZ, and FA, respectively). No private alleles were detected in the pet shop populations.
Genetic diversity varied among wild and pet shop populations (Table 3). The lowest genetic diversity was recorded from the artificial reservoir RS (A = 3.0; AR = 2.6) and the retired fish pond GH (A = 4.0; AR = 3.7). High levels of genetic diversity were found throughout the three per shop populations, with two of them (TZ and FA) reaching the maximum values for A and AR across all sampled populations (Table 3). The highest allelic diversity (A = 5.0; AR = 4.4) in populations from the wild was found in individuals from BW, a chain of man-made ponds used for carp aquaculture which lies southmost of all other wild populations investigated in this study. Private alleles were observed in GH (1), LS (1), and MB (2), all of which varied in level of allelic diversity.
No shop-sourced or wild populations met the requirements for the Hardy–Weinberg Equilibrium (Table 3). The expected heterozygosity (HE) was consistently higher than observed heterozygosity (HO) in all three shop populations and ranged between 0.583 (FA) and 0.613 (TZ and FF). HO ranged between 0.442 (TZ) and 0.496 (FA). Despite a lower sample size in comparison to most other populations, FA also showed the highest HO overall, including wild populations. The lowest observed inbreeding coefficient was observed in the FF population (FIS = 0.152), which was also lowest overall, including wild populations.
In the wild populations, HE ranged between 0.347 (RS) to 0.622 (BW) and HO ranged between 0.274 (RS) and 0.484 (BW). These values were generally lower than those observed in the shop populations, which ranged from Ho = 0.442 to 0.446. Only BW exhibited a level of HO comparable to that of shop populations (HO = 0.484). The lowest inbreeding coefficient was observed in HT (FIS = 0.196). All other wild populations exhibited an inbreeding coefficient > 0.2, which is similar to the values observed in the shop populations. The proportion of common ancestors within each population ranged from F = 0.12 (LS) to F = 0.49 (RS). Both of these systems vary extremely in size, use type, and age.
The proportion of common ancestors within each population as inferred from the F values of the 2MOD program was low in the shop populations and ranged from F = 0.06 to F = 0.07 in FF, TZ, and FA, respectively. These values were lower than those observed in wild populations (F = 0.12—0.23). The highest F value was observed in the wild GH population (F = 0.23). The highest inbreeding value was observed in TZ (FIS = 0.284).
Genetic differentiation
Primarily, shop populations were genetically similar to each other and to some wild populations (Table 4, Fig. 4). No apparent differentiation could be observed in a pairwise comparison between the FF and FA populations (FST = 0.000) and only a slight differentiation (FST = 0.027) was observed between the FF and TZ populations, which was also confirmed in the Jost’s D values (Table 4).
a Discriminant Analysis of Principal Components (DAPC) using 11 microsatellite loci from 10 populations of S. woodiana with ten principal components and three discriminant functions. Similar colors correspond to similar genetic constitutions of respective individuals and populations. b Genetic composition of individuals from all populations based on the DAPC. Dot color corresponds to DAPC results; similar colors correspond to similar genetic composition
We observed distinct differences in genetic structure among wild populations (Table 4, Fig. 4). The lowest level of differentiation was observed between individuals from BW and WZ (FST = 0.047). These systems vary in use type but are relatively close in proximity and belong to a singular drainage system (Danube). Overlaps were also observed between individuals from LS and MB, which vary in structure, use type, and drainage (Danube and Main/Rhine, respectively). The RS population was consistently genetically different from all other wild and shop-sourced populations. The most differences were observed between individuals from RS and WZ (FST = 0. 376), which are geographically close but vary in structure and belong to separate drainages (Main/Rhine and Danube, respectively).
Some shop populations showed a high similarity to some wild populations, including FF and WZ (pairwise FST = 0.071), LS (FST = 0.062), and HT (FST = 0.0383). This result is also supported by the visualization of the genetic clusters created by the DAPC (Fig. 4).
Discussion
In this study, we identified a high level of genetic similarity across wild and shop-sourced S. woodiana populations in Germany, with highest levels of genetic variability observed in pet shop populations. Wild populations shared a single mitochondrial haplotype (HapE-3) and nuclear microsatellite markers indicated high congruence between shop and wild populations. Only one wild population from an artificial reservoir, RS, was structurally different than all other populations, which may be explained by its colonization history.
HapE-3 is the only genetic strain of S. woodiana currently documented in Europe and our results add German individuals to this group. This result aligns with other studies on European populations of S. woodiana, which have confirmed low nucleotide diversity in comparison to native Asian populations (Soroka and Zdanowski 2001; Soroka 2005; Konečný et al. 2018). Some believe this is due to the invasion trajectory of the HapE-3 group, where a primary, single-point introduction gave rise to all current European populations. Low nucleotide diversity is common in invasive species, especially in recent colonizers such as S. woodiana. Their ability to succeed in new environments despite population bottlenecks is well described as the so-called ‘genetic paradox’ of invasion (Hagenblad et al. 2015). In these cases, phenotypic plasticity can facilitate the colonization of new and diverse habitats. In Europe, many shell morphologies have been recorded for S. woodiana, which may allow the mussel to adapt to a broader host fish spectrum, temperatures, and sediment types (Guarneri et al. 2014; Reichard et al. 2022).
Nuclear microsatellite data provided a higher level of resolution and revealed a high level of genetic similarity between shop-sourced individuals, which indicates that pet shops likely all source their mussels from the same or a similar genetic pool. This result, while possibly reflective of the overall genetic constitution of S. woodiana in Europe, may have further been influenced by the relative proximity of our sampled populations, or the short time frame in which we ordered our shop-sourced mussels. It is possible that the genetic relationships between wild and shop populations may be more complex at larger scales or at different points of time.
Shop-sourced individuals were also genetically similar to most wild populations. Currently, information about the collection, distribution, and sale of S. woodiana is very limited. This is true for pet shop websites (pers. obs.) but also literature on the subject (Schoolmann et al. 2006; Patoka et al. 2017; Dobler et al. 2022), making genetic studies an interesting (and sometimes the only) tool to resolve population histories. Due to restrictions which exist regarding the sale of S. woodiana in Europe, it is possible but unlikely that pet shops source their mussels from local, wild populations like the ones included in our study. To our knowledge, only one nature-to-shop pathway has been identified, where a shop-sourced individual was traced to a > 30-year naturalized fish pond in northern Poland (Urbańska et al. 2019). Conversely, shop-to-nature pathways are more common and have been observed in Sweden (Von Proschwitz 2008), Germany (Schoolmann et al. 2006), and Poland (Urbańska and Andrzejewski 2019). While our results cannot confirm the directionality of the mussel’s invasion pathway, we believe it is more likely that the collection, sale, and distribution of S. woodiana by larger suppliers contributes to the admixture of smaller, wild populations. This theory is also supported by the observation that genetic variability was generally greatest in the pet shop populations, and slightly lower in most of the wild populations. Notably, mussels from the Rothsee reservoir were genetically different from all other populations and exhibited the lowest allelic diversity in all populations included in this study. The Rothsee is a large, man-made reservoir that stores water from the Danube River by way of the Main-Danube channel connecting two formerly separate main drainage systems of Main and Danube, before it is released to lower lakes and rivers. The reservoir is a young system that is regularly managed by draining events. Because of this, mussel populations are well monitored in the reservoir, and S. woodiana was first detected there only recently in 2019 (pers. comm; A. Dobler). In this system, most mussels were found close to the shore (max. 4 m depth) and to the in-/outflow of the Main-Danube channel, where they are believed to have been deposited via infested fishes in a historically invaded system (Paunovic et al. 2006; Popa et al. 2011; Lajtner and Crnčan 2011; Dobler et al 2022). The Danube is a major invasion pathway for S. woodiana in Europe. First populations in Croatia were reported in the Danube Basins in Slovakia (Halgoš 1999), Hungary (Vituki 2001), Serbia (Paunovic et al. 2006), and later in Croatia (Graf et al. 2008), where migrating populations ultimately spread into inland lakes and rivers. This natural dispersal mode represents another important source of admixture for European populations. It is possible that the genetic differences observed in the Rothsee population are due to its Danube origin, where its dispersal route has not been subjected to the level of admixture observed in our other inland study sites. The structural uniqueness of the Rothsee population suggests that the dispersal mode of S. woodiana (natural vs anthropogenic spread) may influence its population dynamics.
The results of our study have multiple implications for the management of S. woodiana in Europe. Importantly, steps must be taken on a case-by-case basis to limit the further distribution of S. woodiana by pet shops. The sale of invasive species can be regulated by educating shop vendors and pond owners and by pit-tagging and registering invasive organisms. Researchers must continue to work closely with farmers to identify the most feasible methods for controlling the mussel in colonized ponds. This can include caging fishes in transition ponds before they are moved to other water bodies, or checking gills to ensure that carp and stocking fishes are free from glochidia before releasing them into natural waters (Dobler et al. 2022). Importantly, fish farmers, as well as anglers with whom farmers closely work, must be made aware of the effects that S. woodiana has on native mussel diversity to maximize the impact of management efforts (Urbańska et al. 2021).
Conclusions
The findings of this study contribute to a better understanding of the genetic relatedness and invasion pathways of the non-native Chinese pond mussel in Europe. The close genetic relatedness of pet shop sources as well as most wild populations point at a single source population from which mussels enter the wild. Conversely, the observation of a differentiation of one wild population from all others indicates the occurrence of other dispersal pathways. In almost all cases, comparatively high levels of genetic variability within populations suggest multiple founders and the absence of strong genetic bottlenecks, matching existing knowledge on the great adaptive potential of the species to a range of environmental conditions in newly colonized habitats (Dobler et al., 2022). From a management perspective, preventing the further uncontrolled sale, introduction and dispersal of the species in Germany should become a key priority given its adverse effects on native fauna (Geist et al. 2023).
Data availability
Data will be made available on reasonable request to the corresponding author.
References
Andrzejewski, W., M. Urbańska, J. Mazurkiewicz, H. Gierszal & J. Golski, 2013. The current invasion status of Anodonta woodiana (Lea, 1934) in Poland—study of habitat parameters. Oceanological and Hydrobiological Studies 42(2): 173–180. https://doi.org/10.2478/s13545-013-0071-1.
Bahr, A. L. & V. Wiese, 2018. Freilandvorkommen von Sinanodonta woodiana (Lea 1834) in Ostholstein und Trockenfallen eines Teiches mit umfangreichen Verlusten von Großmuscheln (Bivalvia: Unionidae). Schr Malakozool 30: 39–44.
Benedict, A. & J. Geist, 2021. Effects of water temperature on glochidium viability of Unio crassus and Sinanodonta woodiana: implications for conservation, management and captive breeding. Journal of Molluscan Studies. https://doi.org/10.1093/mollus/eyab011.
Bielen, A., I. Bošnjak, K. Sepčić, M. Jaklič, M. Cvitanić, J. Lušić, J. Lajtner, T. Simčič & S. Hudina, 2016. Differences in tolerance to anthropogenic stress between invasive and native bivalves. Science of the Total Environment 543: 449–459. https://doi.org/10.1016/j.scitotenv.2015.11.049.
Bolotov, I. N., Y. V. Bespalaya, M. Y. Gofarov, A. V. Kondakov, E. S. Konopleva & I. V. Vikhrev, 2016. Spreading of the Chinese pond mussel, Sinanodonta woodiana, across Wallacea: One or more lineages invade tropical islands and Europe. Biochemical Systematics and Ecology 67: 58–64. https://doi.org/10.1016/j.bse.2016.05.018.
Brämick U (2019) Jahresbericht zur Deutschen Binnenfischerei und Binnenaquakultur 2019.
Chapuis, M. P. & A. Estoup, 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24(3): 621–631. https://doi.org/10.1093/molbev/msl191.
Ciofi, C. & M. W. Bruford, 1999. Genetic structure and gene flow among Komodo dragon populations inferred by microsatellite loci analysis. Molecular Ecology 8: S17–S30. https://doi.org/10.1046/j.1365-294X.1999.00734.x.
Dobler, A. H., P. Hoos & J. Geist, 2022. Distribution and potential impacts of non-native Chinese pond mussels Sinanodonta woodiana (Lea, 1834) in Bavaria, Germany. Biological Invasions 24(6): 1689–1706. https://doi.org/10.1007/s10530-022-02737-2.
Donrovich, S. W., K. Douda, V. Plechingerová, K. Rylková, P. Horký, O. Slavík, L. Huan-Zhang, M. Reichard, M. Lopes-Lima & R. Sousa, 2017. Invasive Chinese pond mussel Sinanodonta woodiana threatens native mussel reproduction by inducing cross-resistance of host fish. Aquatic Conservation: Marine and Freshwater Ecosystems 27(6): 1325–1333. https://doi.org/10.1002/aqc.2759.
Douda, K. & Z. Čadková, 2018. Water clearance efficiency indicates potential filter-feeding interactions between invasive Sinanodonta woodiana and native freshwater mussels. Biological Invasions 20(5): 1093–1098. https://doi.org/10.1007/s10530-017-1615-x.
Dudgeon, D. & B. Morton, 1983. The population dynamics and sexual strategy of Anodonta woodiana (Bivalvia: unionacea) in Plover Cove Reservoir, Hong Kong. Journal of Zoology 201(2): 161–183. https://doi.org/10.1111/j.1469-7998.1983.tb04268.x.
EU Regulation No 1143/2014 of 22 October 2014 on the prevention and management of the introduction and spread of invasive alien species.
EU. https://ec.europa.eu/environment/nature/invasivealien/list/index_en.htm (2019).
Gabka, M., Dolata, P. T., & Antonowicz, R. (2007). New localities of the Chinese clam Sinanodonta woodiana (Lea, 1834) (Bivalvia, Unionidae) in the Barycz river valley (Wielkopolska Region). Folia Malacologica. https://doi.org/10.12657/folmal.015.008
Geist, J., 2011. Integrative freshwater ecology and biodiversity conservation. Ecological Indicators 11(6): 1507–1516. https://doi.org/10.1016/j.ecolind.2011.04.002.
Geist, J. & R. Kuehn, 2005. Genetic diversity and differentiation of central European freshwater pearl mussel (Margaritifera margaritifera L.) populations: implications for conservation and management. Molecular Ecology 14(2): 425–439. https://doi.org/10.1111/j.1365-294X.2004.02420.x.
Geist, J., A. Benedict, A. H. Dobler, R. Hoess & P. Hoos, 2023. Functional interactions of non-native aquatic fauna with European freshwater bivalves: implications for management. Hydrobiologia. https://doi.org/10.1007/s10750-022-05121-2.
Goudet, J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Updated from Goudet (1995).
Graf W, Csány B, Leitner P, Paunovic M, Chiriac G, Stubauer I, Ofenböck T, Wagner F (2008) Joint Danube Survey 2 - Final Scientific Report. ICPDR – International Commission for the Protection of the Danube River, http://www.icpdr.org
Guarneri, I., O. P. Popa, L. Gola, L. Kamburska, R. Lauceri, M. Lopes-Lima, L. O. Popa & N. Riccardi, 2014. A morphometric and genetic comparison of Sinanodonta woodiana (Lea, 1834) populations: does shape really matter? Aquatic Invasions. https://doi.org/10.3391/ai.2014.9.2.07.
Hagenblad, J., J. Hülskötter, K. P. Acharya, J. Brunet, O. Chabrerie, Dar Cousins & P.A., Diekmann, M., De Frenne, P., Hermy, M., Jamoneau, A., Kolb, A., Lemke, I., Plue, J., Reshi, Z.A., S. A., & Graae, B. J., 2015. Low genetic diversity despite multiple introductions of the invasive plant species Impatiens glandulifera in Europe. BMC Genetics 16(1): 1–16. https://doi.org/10.1186/s12863-015-0242-8.
Halgoš, 1999. Mass occurrence of Anodonta woodiana (Lea, 1834) in Slovakia. Folia Faunistica Slovaca 4: 7.
Huber, V. & J. Geist, 2019. Reproduction success of the invasive Sinanodonta woodiana (Lea 1834) in relation to native mussel species. Biological Invasions 21(11): 3451–3465. https://doi.org/10.1007/s10530-019-02060-3.
Hulme, P. E., S. Bacher, M. Kenis, S. Klotz, I. Kühn, D. Minchin, W. Nentwig, S. Olenin, V. Panov, J. Pergl, P. Pyšek, A. Roques, D. Sol, W. Solarz & M. Vilà, 2008. Grasping at the routes of biological invasions: a framework for integrating pathways into policy. Journal of Applied Ecology 45(2): 403–414. https://doi.org/10.1111/j.1365-2664.2007.01442.x.
Jombart, T., S. Devillard & F. Balloux, 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11(1): 1–15. https://doi.org/10.1186/1471-2156-11-94.
Jost, L., 2008. GST and its relatives do not measure differentiation. Molecular Ecology 17(18): 4015–4026.
Karaouzas, I., D. V. Gonçalves, N. Riccardi, S. Varandas, E. Froufe, S. Zogaris & M. Lopes-Lima, 2022. First established population of the Chinese pond mussel Sinanodonta woodiana (Lea, 1834) (Bivalvia: Unionidae) in Greece. BioInvasions Record 11(1): 110. https://doi.org/10.3391/bir.2022.11.1.12.
Kondakov, A. V., Y. V. Bespalaya, I. V. Vikhrev, E. S. Konopleva, M. Y. Gofarov, A. A. Tomilova, V. V. Maxim & I. N. Bolotov, 2020. The Asian pond mussels rapidly colonize Russia: successful invasions of two cryptic species to the Volga and Ob rivers. BioInvasions Records 9(3): 504–518. https://doi.org/10.3391/bir.2020.9.3.07.
Konečný, A., O. P. Popa, V. Bartáková, K. Douda, J. Bryja, C. Smith, L. O. Popa & M. Reichard, 2018. Modeling the invasion history of Sinanodonta woodiana in Europe: Tracking the routes of a sedentary aquatic invader with mobile parasitic larvae. Evolutionary Applications 11(10): 1975–1989. https://doi.org/10.1111/eva.12700.
Labecka, A. M. & J. Domagala, 2018. Continuous reproduction of Sinanodonta woodiana (Lea, 1824) females: an invasive mussel species in a female-biased population. Hydrobiologia 810(1): 57–76. https://doi.org/10.1007/s10750-016-2835-2.
Labecka, A. M. & M. Czarnoleski, 2021. Patterns of growth, brooding and offspring size in the invasive mussel Sinanodonta woodiana (Lea, 1834) (Bivalvia: Unionidae) from an anthropogenic heat island. Hydrobiologia 848(12): 3093–3113. https://doi.org/10.1007/s10750-019-04141-9.
Lajtner, J. & P. Crnčan, 2011. Distribution of the invasive bivalve Sinanodonta woodiana (Lea, 1834) in Croatia. Aquatic Invasions 6(1): 119–124. https://doi.org/10.3391/ai.2011.6.S1.027.
Light, T., & Moyle, P. (2015). 19. Assembly Rules and Novel Assemblages in Aquatic Ecosystems. Biological Invasions in Changing Ecosystems. (Ed. J. Canning-Clode), De Gruyter Open, Warsaw, Poland. https://doi.org/10.1049/el.2015.0684
Najberek, K., M. Strzalka & W. Solarz, 2011. Alien Sinanodonta woodiana (Lea, 1834) and protected Anodonta cygnea (Linnaeus, 1758) (Bivalvia: Unionidae) in the Spytkowice pond complex. Folia Malacologica. https://doi.org/10.2478/v10125-011-0011-x.
Nunes, A. L., E. Tricarico, V. E. Panov, A. C. Cardoso & S. Katsanevakis, 2015. Pathways and Gateways of Freshwater Invasions in Europe. https://doi.org/10.3391/ai.2015.10.4.01.
Patoka, J., M. Bláha, L. Kalous & A. Kouba, 2017. Irresponsible vendors: Non-native, invasive and threatened animals offered for garden pond stocking. Aquatic Conservation: Marine and Freshwater Ecosystems 27(3): 692–697. https://doi.org/10.1002/aqc.2719.
Paunovic, M., B. Csányi, V. Simic, B. Stojanovic & P. Cakic, 2006. Distribution of Anodonta (Sinanodonta) woodiana (Rea, 1834) in inland waters of Serbia. Aquatic Invasions 1(3): 154–160. https://doi.org/10.3391/ai.2006.1.3.10.
Peakall, R. & P. E. Smouse, 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. https://doi.org/10.1093/bioinformatics/bts460.
Popa, O. P., L. O. Popa, A. M. Krapal, D. Murariu, E. I. Iorgu & M. Costache, 2011. Sinanodonta woodiana (Mollusca: Bivalvia: Unionidae): Isolation and characterization of the first microsatellite markers. International Journal of Molecular Sciences 12(8): 5255–5260. https://doi.org/10.3390/ijms12085255.
Popa, O. P., V. Bartáková, J. Bryja, M. Reichard & L. O. Popa, 2015. Characterization of nine microsatellite markers and development of multiplex PCRs for the Chinese huge mussel Anodonta (Sinanodonta) woodiana Lea, 1834 (Mollusca, Bivalvia). Biochem Syst Ecol 60(23): 234. https://doi.org/10.1016/j.bse.2015.05.001.
Poznańska-Kakareko, M., K. Wiśniewski, D. Szarmach, A. Witkowska, T. Kakareko & J. Kobak, 2021. Importance of substratum quality for potential competitive niche overlap between native and invasive unionid mussels in Europe. Science of the Total Environment 799: 149345. https://doi.org/10.1016/j.scitotenv.2021.149345.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155(2): 945–959. https://doi.org/10.1093/genetics/155.2.945.
R Core Team (2022). R: A language and environment for statistical ## computing. R Foundation for Statistical Computing, Vienna, Austria.
Raymond, M. & F. Rousset, 1995. An exact test for population differentiation. Evolution. https://doi.org/10.1111/j.1558-5646.1995.tb04456.x.
Reichard, M., K. Douda, R. Blažek & A. Janovská, 2022. Increased plasma cortisol level as acute response to glochidia parasitism. Environmental Biology of Fishes. https://doi.org/10.1007/s10641-022-01379-6.
Rousset, F., 2008. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8(1): 103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x.
Sárkány-Kiss, A., I. Sîrbu & O. Hulea, 2000. Expansion of the adventive species Anodonta woodiana (Lea, 1834) (Mollusca, Bivalvia, Unionoidea) in central and eastern Europe. Acta Oecol Univ Sibiu 7: 49–57.
Schoolmann, G., A. Martens & K. Grabow, 2006. Einschleppung und Verbreitung der Chinesischen Teichmuschel Sinanodonta woodiana (Lea) durch den Zoo-und Gartenfachhandel (Bivalvia: Unionidae). Lauterbornia 58: 139–141.
Soroka, M., 2005. Genetic Variability Among Freshwater Mussel Anodonta woodiana (Lea, 1834) (Bivalvia: Unionidae) Populations Recently Introduced in Poland. Zoological Science 22(10): 1137–1144. https://doi.org/10.2108/zsj.22.1137.
Soroka, M. & B. Zdanowski, 2001. Morphological and genetic variability of the population of the population of Anodonta woodiana (Lea, 1834) occurring in the heated Konin lakes system. Arch. Pol. Fish. 9: 239–252. https://doi.org/10.2108/zsj.22.1137.
Soroka, M., M. Urbańska & W. Andrzejewski, 2014. Chinese pond mussel Sinanodonta woodiana (Lea, 1834) (Bivalvia): origin of the Polish population and GenBank data. Journal of Limnology. https://doi.org/10.4081/jlimnol.2014.938.
Sousa, R., A. Novais, R. Costa & D. Strayer, 2014. Invasive bivalves in fresh waters: impacts from individuals to ecosystems and possible control strategies. Hydrobiologia 735: 233–251. https://doi.org/10.1007/s10750-012-1409-1.
Spyra, A., N. Jędraszewska, M. Strzelec & M. Krodkiewska, 2016. Further expansion of the invasive mussel Sinanodonta woodiana (Lea, 1834) in Poland–establishment of a new locality and population features. Knowledge & Management of Aquatic Ecosystems 417: 41. https://doi.org/10.1051/kmae/2016028.
Strayer, D. L., J. Powell, P. Ambrose, L. C. Smith, M. L. Pace & D. T. Fischer, 1996. Arrival, spread, and early dynamics of a zebra mussel (Dreissena polymorpha) population in the Hudson River estuary. Canadian Journal of Fisheries and Aquatic Sciences 53(5): 1143–1149. https://doi.org/10.1139/f96-038.
Urbańska, M., & Andrzejewski, W. (2019). An invasion in progress-Sinanodonta woodiana (Lea, 1834) (Bivalvia: Unionidae) in Poland. Folia Malacologica, 27(4), 327–335. https://doi.org/10.12657/folmal.027.022
Urbańska, M., M. Kirschenstein, K. Obolewski & M. Ożgo, 2019. Silent invasion: Sinanodonta woodiana successfully reproduces and possibly endangers native mussels in the north of its invasive range in Europe. International Review of Hydrobiology 104(5–6): 127–136. https://doi.org/10.1002/iroh.201801971.
Urbańska, M., A. Kamocki, M. Kirschenstein & M. Ożgo, 2021. The Chinese pond mussel Sinanodonta woodiana demographically outperforms European native mussels. Scientific Reports 11(1): 1–11. https://doi.org/10.1038/s41598-021-96568-1.
Van den Neucker, T. & K. Scheers, 2022. Mislabelling may explain why some prohibited invasive aquatic plants are still being sold in Belgium. Knowledge & Management of Aquatic Ecosystems 423: 8. https://doi.org/10.1051/kmae/2022005.
Van Oosterhout, C., W. F. Hutchinson, D. P. Wills & P. Shipley, 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4(3): 535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x.
Vituki (2001) Second phase of surveying the environmental and natural damages, which were caused by the cyanide and heavy metal pollution accidents of Romanian sources in the Szamos and Tisza river systems. Final Report 712/3/488902, Institute for Water Pollution Control, Budapest. p. 43
Wachtler, K., M. C. Dreher-Mansur & T. Richter, 2001. Larval types and early postlarval biology in naiads (Unionoida), Bauer G Wachtler K. Ecology and evolution of the freshwater mussels Unionoida. Springer, Berlin: 93–125.
Weir, B. S. & C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Zieritz, A., B. Gum, R. Kuehn & J. Geist, 2012. Identifying freshwater mussels (Unionoida) and parasitic glochidia larvae from host fish gills: a molecular key to the North and Central European species. Ecology and Evolution 2(4): 740–750. https://doi.org/10.1002/ece3.220.
Acknowledgements
We particularly thank Andreas Dobler and Philipp Hoos of the Muschelkoordinationstelle Bayern for their help in conceiving this project and for collecting mussels used in this study. We also thank Christine Seidel, Elke Siegel, and Sahin Gündüz for their help with laboratory work as well as all fish pond owners for their kind cooperation.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
This study was conceived by JG, RK, BS, AB, and Andreas Dobler and Philipp Hoos of the Muschelkoordinationstelle Bayern. Mussels were collected by AB with help from Andreas Dobler and Philipp Hoos. Genotyping and microsatellite data analysis was performed by AB with help from BS. The original manuscript was drafted by AB with help from JG. All authors contributed to the review and editing and read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors have no financial interests to disclose.
Additional information
Handling editor: Diego Fontaneto
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Benedict, A., Kuehn, R., Stoeckle, B.C. et al. Genetic comparisons of the invasive pond mussel Sinanodonta woodiana from wild and pet shop populations in Germany. Hydrobiologia 851, 2125–2137 (2024). https://doi.org/10.1007/s10750-023-05442-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05442-w