Abstract
Habitat fragmentation is among the main threats to freshwater fish biodiversity, with expected effects including genetic impacts due to disturbance of migration and declining population size. Major concern falls on highly exploited species such as Prochilodus magdalenae, a migratory characiform fish endemic to the Magdalena River basin supporting much of the Colombian artisanal fishery, whose migration route was interrupted by the Ituango hydroelectric project in the Cauca River. To determine the potential effects of fragmentation, this study evaluated the population genetics of this species on both geographical (upstream and downstream of the dam) and temporal (before and after the construction) scales by using 11 species-specific microsatellite markers. Contrary to expectation, genetic diversity and structure remained relatively stable. This study provided no evidence of genetic impacts on P. magdalenae associated with fragmentation over the short term (4–10 years, 5–12 generations) despite persisting threats, genetic evidence of bottleneck, and a high degree of inbreeding, showing the ability of this species to withstand disturbance of its habitat.
Similar content being viewed by others
Introduction
The global abundance of freshwater faunal stocks decreased by 84% between 1970 and 2016 (WWF, 2020), mainly due to overexploitation, water pollution, habitat degradation, water-flow modification, and species invasion (Dudgeon et al., 2006), in addition to climate change and the increase in emerging pollutants (Arthington et al., 2016; Reid et al., 2019). One of the most important disturbances in freshwater systems is the construction of dams, including disruption of the life cycle of migratory species (Liermann et al., 2012; Reis et al., 2016; Barbarossa et al., 2020; Herrera-Pérez et al., 2019; Reid et al., 2019). It is estimated that more than 76% of migratory river fish populations have been reduced since 1970 (Deinet et al., 2020), while construction of dams has accelerated and could affect almost 93% of the rivers around the planet by 2030 (Grill et al., 2015, 2019).
The Magdalena-Cauca basin, located in the northern Andes Mountains, concentrates most of the Colombian human population and is a key area for the economy of the country (Restrepo et al., 2020). It has high aquatic biodiversity, with more than 235 fish species, and supports a large part of the Colombian artisanal fishery, with more than 14,300 tons landed in 2019 (García-Alzate et al., 2020; Valderrama et al., 2020). However, the biodiversity of this basin is threatened, including 54 of said species classified with some degree of vulnerability in the local red list (Mojica et al., 2012). These species are impacted by various anthropogenic activities related to water pollution, mining, livestock production, resource overexploitation, and habitat modification, including construction of dams (López-Casas et al., 2020).
One of these species is the characiform fish bocachico Prochilodus magdalenae Steindachner 1879, the most important fishery resource, with roughly 40% of total landings of the Magdalena-Cauca River basin in 2019 (Duarte et al., 2019; Valderrama et al., 2020), albeit with an estimated reduction of 76% of their fishery production since 1975 (Barreto, 2017). This species was classified as Critically Endangered in 2002 and has been listed as Vulnerable since 2012, in the Colombian red list of threatened freshwater fishes (Mojica et al., 2012). The national authorities have established a minimum landing size of 25 cm for this detritivorous fish, which inhabits the Magdalena, Atrato, and Sinú basins. This fish reaches sexual maturity at an average size of 20–25 cm and between 1 and 1.5 years of age, and can attain a maximum standard length of nearly 50 cm (Lasso et al., 2011). This species is iteroparous, meaning that it reproduces multiple times throughout its lifespan, and is known for its massive spawning events (Jiménez-Segura et al., 2010). This fish undergoes long longitudinal migrations, with reports of distances of up to 1,223 km (López-Casas et al., 2016), and exhibits two migration peaks per year that coincide with the flood cycles of the Cauca and Magdalena rivers (Barletta et al., 2015; Jiménez-Segura et al., 2010, 2016).
The population genetics of P. magdalenae has been more studied than for other fish species of the Magdalena-Cauca River basin (Márquez et al., 2020). The use of microsatellite DNA markers demonstrated the presence of populations with high genetic diversity, a significant degree of inbreeding, and the coexistence of two main genetic groups (stocks) in the Magdalena basin, as well as genetic differentiation among populations from the Magdalena, Atrato and Sinú basins (Márquez et al., 2020). Additionally, despite the river connectivity and the wide native distribution of this species, the unexpected findings of spatial genetic structure within the Magdalena Basin may be explained by hybridization with related species and anthropogenic effects on its habitat such as restocking and fish translocation (Fontalvo et al., 2018; Orozco-Berdugo & Narváez-Barandica, 2014; Landínez-García et al., 2020). Although national and regional entities have made attempts to restock wild populations of this species in the last 20 years (see discussion in Landínez-García et al., 2020), detailed information of such practices remains unavailable.
The increasing environmental threats to freshwater fish species demand genetic evaluation of wild populations to assess microevolutionary processes that reflect landscape changes, infer the adaptive evolutionary risks and inform management and conservation planning (Manel et al., 2003; Schwartz et al., 2007; Laikre et al., 2010; Epps & Keyghobadi, 2015). While empirical studies about anthropogenic impacts on freshwater fish genetics are limited, impacts of isolation by dams on migratory fish populations at different geographic scales are documented (e.g.,Yamamoto et al., 2004; Van Leeuwen et al., 2018; Klütsch et al., 2019; Ruzich et al., 2019; Liu et al., 2020; Pimentel et al., 2020; Vega-Retter et al., 2020). In the Neotropics, studies of migratory fish populations fragmented by dams reported differences in genetic diversity and structure between upstream and downstream sections for species such as Pseudoplatystoma corruscans (Spix & Agassiz 1829) (Paraná River, Brazil; Prado et al., 2018), Prochilodus magdalenae (Magdalena River, Colombia; Fontalvo et al., 2018), Prochilodus costatus Valenciennes 1850 (San Francisco River, Brazil; Pimentel et al., 2020), Trichomycterus areolatus (Valenciennes 1846), and Basilichthys microlepidotus (Jenyns 1841) (Choapa River, Chile; Vega-Retter et al., 2020). This last study found significant changes on a temporal scale in the short term (5 years) after dam construction, whereas species such as Pimelodus maculatus Lacepède 1803 in the Upper Uruguay River basin (Ribolli et al., 2012) and Prochilodus lineatus (Valenciennes 1837) in the Paraná River (Ferreira et al., 2017) showed gene flow and high genetic diversity after similar (2–10 years) and even longer (15–30 years) periods of fragmentation, respectively. Indeed, the detection of genetic differences resulting from the fragmentation of rivers and the level of impact seem to depend upon complex interactions of factors that include the degree of fragmentation and time elapsed since the disturbance (Ruzich et al., 2019; Liu et al., 2020; Vega-Retter et al., 2020), presence, or absence of fishways that facilitate migration (Ferreira et al., 2017) or translocation and restocking of fish (Klütsch et al., 2019; Pimentel et al., 2020), generation interval of each species (Epps & Keyghobadi, 2015; Ruzich et al., 2019), and pre-existing genetic diversity and structure (Epps & Keyghobadi, 2015; Landguth et al., 2010; Coleman et al., 2018).
The Cauca River is 1,350 km long and runs from south to north between the western and central Andes Mountains, from Laguna del Buey (Cauca Department) to Pinillos (Bolívar Department) in the Colombian Caribbean basin (Rodríguez-Olarte et al., 2011). The upper part of the river predominantly meanders, descending to about 900 m above sea level, while the middle and lower parts flow through a steep, narrow valley over the last 500 km, from the confluence with the Risaralda River to its mouth in the Magdalena River. This stretch of the river is known for the rapids and waterfalls that function as natural barriers to several migratory fish species (Galvis & Mojica, 2007; Rodríguez-Olarte et al., 2011), and it is where the Ituango Dam is located. The Ituango Dam is the largest in Colombia, standing 225 m high with a crest that is 550 m long and has no fishways. Its construction began in 2010, and it is expected to produce 2400 MW of power per year.
Given the recent fragmentation of populations of P. magdalenae by the Ituango hydroelectric project dam since 2014, persisting threats to its habitat, and the knowledge gap about short-term effects of these factors on fishes in the Cauca River, this study evaluated the population genetics of P. magdalenae collected between 2019 and 2021 in eight sections of the middle and lower Cauca River after the fragmentation (Ex-post sample). Thus, this study evaluates the hypotheses of both spatial (upstream–downstream of the dam) and temporal genetic differences (before–after dam construction) in P. magdalenae of the Cauca River over a period of 4–10 years. These hypotheses are based on (i) the genetic vulnerability of populations of this species in the Cauca River, showing genetic evidence of inbreeding and genetic bottleneck before the fragmentation (Landínez-García et al., 2020), (ii) the potential disruption of gene flow, (iii) likely effective population size changes between upstream and downstream populations due to the fragmentation as has occurred in other species (Pavlova et al., 2017), (iv) the passage of several generations after the fragmentation event, allowing the detection of potential changes (Tallmon et al., 2010; Epps & Keyghobadi, 2015), and (v) the likelihood that the genetic diversity of high-dispersal species may reduce the time lag required to detect the potential impacts of habitat perturbations (Landguth et al., 2010; Fluker et al., 2014; Epps & Keyghobadi, 2015). Therefore, this study can contribute to understanding the dynamic relation between population genetics of the freshwater fish fauna and habitat disturbances.
Materials and methods
Sampling and genotyping
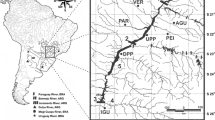
A total of 384 muscle or fin tissue samples from Prochilodus magdalenae were collected between 2019 and 2021 (Ex-post sample) from sections of the middle and lower parts of the Cauca River upstream (S1, PHI) and downstream (S2, S3, S4, S5, S6, and S7/S8) of the Ituango Dam (Fig. 1). The sections referred to in this study were previously sampled by Landínez-García and Márquez (2016) and Landínez-García et al. (2020) and were used for comparison with historical sample data (Ex-ante sample). PHI, a stretch of the river known for the presence of rapids and waterfalls, is the section where the Ituango Dam was constructed. Samples preserved in 70% ethanol were supplied by the Grupo de Ictiología from the Universidad de Antioquia (GIUA), Fundación Humedales, and Grupo de Biotecnología Animal from Universidad Nacional de Colombia, Sede Medellín. The sample sizes collected Ex-ante (Landínez-García et al., 2020) and Ex-post (this study) analyzed in this research were as follows: S1: 33|37, PHI: 0|25, S2: 30|18, S3: 28|13, S4: 38|141, S5: 40|28, S6: 94|36, S7/S8: 45|52.
Sampling points (triangles) for Prochilodus magdalenae after the construction (Ex-post sample) of the Ituango Dam grouped into eight sections of the Cauca River, Colombia. S1 Bolombolo, Puente Real, PHI Barbacoas, Brugo, Orobajo, La Pená, San Cristóbal Pená, Peque, Santa María, Sardinas, Llano Grande, S2 El Aro, Gurimán, La Guamera, S3 Ituango River mouth, Espíritu Santo River mouth, S4 Cáceres, Caucasia, Tarazá, S5 Barrio Chino, Palanca, floodplains La Ilusión and Palomar, S6 floodplains Grande, La Caimanera and La Raya, S7/S8 Tres Cruces, floodplains El Floral, Nueva and La Panela
DNA extraction and PCR amplification of 11 species-specific microsatellite loci (Pma01, Pma02, Pma13, Pma14, Pma18, Pma25, Pma35, Pma36, Pma39, Pma40, and Pma46) were performed according to Landínez-García et al. (2020). The amplified fragments were separated by capillary electrophoresis in an ABI 3730 XL automated DNA sequencer (Applied Biosystems), using LIZ600 (Applied Biosystems) as an internal molecular size marker. Alleles were scored in GeneMarker software v.3.0.0, and amplification, scoring, and genotyping errors were evaluated using Micro-Checker v.2.2.3 (Van Oosterhout et al., 2004).
Genetic diversity and outlier loci screening
To quantify the genetic diversity of P. magdalenae in eight sections of the Cauca River from the Ex-post sample, the average value per locus of the number of alleles (Na), allelic size range (Ra), and expected (HE) and observed (HO) heterozygosities were calculated using GenAlEx v6.51b2 (Peakall & Smouse, 2006, 2012). The allelic richness (Ar), for comparison of the number of alleles accounting for differing sample sizes, was calculated in FSTAT v.2.9.4 (Goudet, 2003). Inbreeding coefficients (FIS) and deviations from Hardy–Weinberg (HWE) and linkage (LD) equilibria were evaluated in Arlequin v3.5.2.2 (Excoffier & Lischer, 2010). Fisher's Exact Test, implemented in the web version of Genepop v4.7.5 (Raymond & Rousset, 1995; Rousset, 2008), allowed calculation of the multilocus statistical significance of departures from HWE for collections from each river section or population. Bar plots were used for descriptive comparison among the genetic diversity metrics (Na, Ar, Ra, HE, HO, and FIS) with those reported by Landínez-García et al. (2020) in the Ex-ante sample (308 individuals, years 2010–2014). Additionally, the G-test (modified Fisher's exact probability test) implemented in the web version of Genepop v4.7.5 (Raymond & Rousset, 1995; Rousset, 2008) was used to evaluate the null hypothesis that the alleles from the Ex-ante and Ex-post samples were taken from the same distribution.
To identify candidate loci under natural selection (outlier loci) in the Ex-post sample, the BayeScan v2.1 software (Foll & Gaggiotti, 2008) was used setting 10:1 as the prior odds for the neutral model with respect to the model with selection, with 20 pilot runs and 5,000 iterations each, and a total of 250,000 subsequent iterations with 50,000 as the burn-in period. Loci with a posterior probability (PP) > 0.75 were considered significant. Samples with outlier loci were excluded from subsequent population genetic analyses, and the genetic diversity estimators were recalculated.
Demographic estimations
Two methods were applied to detect drastic reduction in population size or recent bottlenecks in the population. The first method involved calculating the standardized M index of Garza & Williamson (2001) using Arlequin v3.5.2.2 (Excoffier & Lischer, 2010). A reduction in the number of alleles with respect to the allelic size range of a population may indicate recent bottleneck events. The second method evaluated excess heterozygosity using the software BOTTLENECK v1.2.02 (Piry et al., 1999) with default settings and three mutation models of microsatellites in the coalescent simulation (Infinite Alleles Model—IAM, Two-Phase Model—TPM, Stepwise Mutation Model—SMM; Cornuet & Luikart, 1996; Luikart & Cornuet, 1998). The Bonferroni correction was applied in cases of multiple comparisons.
Estimation of the effective population size (Ne) was performed in NeEstimator v2.1 (Do et al., 2014) implementing the linkage disequilibrium (LD) method, which used a point estimation in each Ex-ante and Ex-post sample, ignoring allelic frequencies ≤ 0.02 that could overestimate Ne (Waples & Do, 2010; Do et al., 2014). A second approach included the temporal method, which estimates a single value of Ne from two or more sets of samples separated by n generations (Do et al., 2014). A rough calculation of n for P. magdalenae between the Ex-ante and Ex-post samples was based on a generation length of 1/mortality + age of first reproduction—1 (IUCN Standards and Petitions Committee, 2019), considering a total mortality of 3.45 individuals per year in the Cauca River (Zárate, Valderrama & Atencio, Fundación Humedales, personal communication), an average age of first maturity at 1.5 years (Lasso et al., 2011), and an average time between samples of seven years. The other parameters considered in the temporal method were the estimator Fs, which shows less bias than its analogs Fc and Fk when evaluating the change in allele frequencies (Jorde & Ryman, 2007), jackknife confidence intervals (CIs), and sampling method Plan II, which does not require knowing the population size (N) to perform the estimation (Do et al., 2014). When Ne estimates yielded infinite values, this was interpreted as insufficient evidence of a finite population size, as the larger sampling variance compared to genetic drift signals suggests that the data cannot reliably support estimation of a finite Ne (Waples & Do, 2010).
The Geneclass2 software (Piry et al., 2004) was used to assess interpopulation migration events for the Ex-post sample, for which, based on the sampled sections, each of the individuals was assigned to its most probable section of origin. This approach used a Bayesian criterion using the maximum likelihood estimator L_home/L_max (relationship between the maximum likelihood of the individual of having originated from the sampled section and the maximum likelihood among all sections) and the statistical Monte Carlo resampling method proposed by Paetkau et al. (2004), with 10,000 individuals simulated and an error probability of 0.01.
Genetic structure
Analysis of molecular variance (AMOVA; Meirmans, 2006) and calculation of standardized pairwise indices of genetic distance F'ST (Meirmans, 2006; Meirmans & Hedrick, 2011) and DEST (Jost, 2008; Meirmans & Hedrick, 2011) using the GenAlEx v6.51b2 (Peakall & Smouse, 2006, 2012), applying the Bonferroni correction for multiple comparisons, were used to explore the genetic structure of the Ex-post sample of P. magdalenae in the Cauca River. Discriminant analysis of principal components (DAPC) implemented in the R package Adegenet (Jombart, 2008) was also employed and allowed visualizing spatial genetic structure by maximizing the differences in multilocus genotypes among collections. Bayesian clustering analysis using Structure v2.3.4 (Pritchard et al., 2000) was further utilized implementing the genetic mixture and correlated alleles models, with 600,000 Monte Carlo Markov chains with 100,000 regarded as the burn-in period. This study evaluated 1 ≤ K ≤ m + 3, where m is the number of a priori populations (Evanno et al., 2005), with 20 repetitions each. The most likely number of populations (K) was determined after comparing five different statistics (MedMeaK, MaxMeaK, MedMedK and MaxMedK: see Puechmaille, 2016; and ΔK: see Evanno et al., 2005) in addition to the histogram of the co-ancestry probabilities to each genetic stock, obtained in the StructureSelector online software (Li & Liu, 2018). A co-ancestry threshold value of 0.5 and 0.7 was applied to cluster the samples to each genetic stock in the Ex-ante and Ex-post samples, respectively. These methods were replicated for comparing the Ex-ante and Ex-post temporal samples for each genetic stock, considering both samples as different populations. Additionally, each temporal sample was divided into upstream and downstream subsamples, performing pairwise comparisons of F´ST and DEST indices to infer genetic differences by fragmentation.
Results
Outlier screening and genetic diversity
A set of 34 samples from the Grande floodplain in section S6 were excluded from the population genetic analysis as they exhibited four outlier loci (Online Resource 1). The eight sections of the Ex-post sample showed similar indices of genetic diversity (Table 1), with average number of alleles per locus in the range of 11.45 (S3)–19.62 (S4), allelic richness of 10.32 (PHI)–11.00 (S5) and expected heterozygosities of 0.885 (PHI)–0.908 (S5). There was no evidence of LD among loci, and each section showed significant departures from HWE and significant inbreeding values (17.4–23.3%; Table 1). In the overall sample, null alleles were detected at 7 loci (Pma18, Pma14, Pma01, Pma36, Pma40, Pma39, and Pma35), with frequencies ranging from 0.10 to 0.18. Two genetic stocks were suggested by some of the genetic structure analyses (stocks are described below in Genetic structure) with differences in genetic diversity indices, and showing similar and significant inbreeding values (Table 2).
All indices of genetic diversity for the Ex-post sample (2019–2021) were comparable to those for the Ex-ante sample (2010–2014; Fig. 2). However, the G-test showed that seven of the 11 loci (Pma18, Pma13, Pma14, Pma40, Pma39, Pma02, Pma25) had temporal changes in the distribution of allele frequencies (P < 0.05).
Mean per-locus genetic diversity metrics for Prochilodus magdalenae collected from eight sections of the Cauca River before (Ex-ante: 2010–2014) and after (Ex-post: 2019–2021) the construction of the Ituango Dam. Na number of alleles, Ar allelic richness, Ra allelic range, HE expected heterozygosity, HO observed heterozygosity, and FIS inbreeding coefficient
Demographic estimations
Results of test to detect recent bottleneck showed M indices in the range of 0.15–0.23 for the nine sections and an overall value of 0.26 (Table 3); all were smaller than the reference value of 0.68 (Garza & Williamson, 2001). Additionally, the heterozygosity excess test results were significant (P < 0.006) under the IAM (all sections and overall). These results present genetic evidence of a recent bottleneck for P. magdalenae of the Cauca River.
The estimated effective population size using the linkage disequilibrium method (LD) was higher in the Ex-post sample (Ne = 7000.8; CI 1,875.7-∞) than in the Ex-ante sample (2875.8, CI 1226.7-∞). In turn, these both values were higher than that obtained in the temporal method (755.3, CI: 423.8–3461.1), which was estimated using an average of n = 7 generations for P. magdalenae between the years 2010 and 2021. Additionally, it was not possible to estimate Ne in each genetic stock as estimations yielded infinite values (Ne = ∞; Table 3).
Finally, in the Ex-post sample, eight individuals were identified as likely migrants (P < 0.01) among fragmented river sections according to the results of the first-generation migrant detection test. Specifically, seven individuals collected downstream of the Ituango dam were detected as likely immigrants from upstream sections, while one individual collected upstream was identified as likely immigrant from downstream sections.
Genetic structure
No significant pairwise genetic differences were detected within the Ex-post sample (350 individuals, years 2019–2021) by the F'ST and Jost's DEST indices (Table 4). The geographic pattern of genetic variation was quantified as variance within individuals (77%), among individuals (23%), and among Sects. (0%), although the overall structure index calculated using the AMOVA was significant (F´ST (7699) = 0.045, P = 0.001). Moreover, the absence of spatial genetic differentiation was graphically evident by observation of overlapping of points representing the respective sections in DAPC (Fig. 3a) and by the mixture of genetic stocks along the sampled area according to the distribution of the co-ancestry probabilities of the Bayesian analysis (Fig. 3b), since K = 2 was the most likely number of populations based on the five statistics calculated: MedMeaK, MaxMeaK, MedMedK, MaxMedK, and ΔK.
a Results of discriminant analysis of principal components (DAPC; 90 principal components retained, explaining 85% of the variance), and b co-ancestry probabilities based on the Bayesian analysis of 350 individuals of Prochilodus magdalenae in the Ex-post sample (2019–2021, sections S1, PHI, S2, S3, S4, S5, S6, and S7/S8 of the Cauca River)
At the temporal scale, comparisons between Ex-ante and Ex-post samples in both stocks showed differences in the AMOVA results and standardized pairwise indices of genetic distances. For stock 1, the results were as follows: F´ST (1, 553) = 0.080, P = 0.001, and DEST = 0.083, P = 0.001. For stock 2, the results were F´ST (1, 545) = 0.106, P = 0.001 and DEST = 0.067, P = 0.001. However, the genetic variation was mostly explained by the variance within individuals (stock 1: 79%, stock 2: 78%) and among individuals (stock 1: 20%, stock 2: 21%), rather than between periods (only 1% in both stocks). Additionally, the temporal samples were not clearly discriminated by the DAPC (Fig. 4a). On the other hand, the fragmented sections affected by the dam showed no genetic differences either over space or time (Table 5).
In the Bayesian analysis, the most likely number of populations was estimated as K = 2 for the MedMeaK, MaxMeaK, MedMedK, and MaxMedK estimators, and K = 4 for ΔK. The histogram of K = 2 showed the presence of a genetic stock mixture throughout the studied sections in the Cauca River (Fig. 4b).
Discussion
This study used temporal samples to compare population genetics of Prochilodus magdalenae before and after construction of a dam in the Cauca River in Colombia to assess genetic impacts attributable to habitat fragmentation. Dam construction often involves water-flow pattern modification and disruption of the dispersal and migration patterns of freshwater fishes, which in turn may impact spatial genetic structure by interruption of gene flow as well as changes in the amount and patterning of genetic variability (Baggio et al., 2018; Tamario et al., 2019). Suitable evaluation of those hypotheses may require temporal approaches that are difficult to achieve in practice, using empirical genetic data to deal with confounding factors related to other habitat disturbances (Schwartz et al., 2007; Epps & Keyghobadi, 2015). Indeed, such studies are few, and results often cannot be generalized due to differing life-history traits of the species or particular habitat conditions (see Beneteau, et al., 2008; Ruzich et al., 2019; Yamamoto et al., 2019; Liu et al., 2020; Vega-Retter et al., 2020).
This study showed no differences in the genetic diversity of P. magdalenae in the Cauca River between fragmented sections of the Ex-post sample (period 2019–2021). Moreover, HE values remained higher than the average for Neotropical characiforms using species-specific microsatellites (HE = 0.675; see Hilsdorf & Hallerman, 2017) and those reported in its congeners Prochilodus argenteus Spix & Agassiz 1829 (HE: 0.690—0.827; Sanches et al., 2012; Melo et al., 2013; Coimbra et al., 2017), P. costatus (HE: 0.490–0.867; Carvalho-Costa et al., 2008; Melo et al., 2013; Pimentel et al., 2020) and P. lineatus (HE: 0.607—0.858; Rueda et al., 2013; Ferreira et al., 2017; Iwersen et al., 2019; Lopera-Barrero et al., 2016, 2019). Such stability in genetic diversity between fragmented sections may be explained not only by large effective population sizes, but also by the small number of generations produced within the short time that elapsed since the fragmentation of the river (Epps & Keyghobadi, 2015; Ruzich et al., 2019). Additionally, no changes were detected with respect to the Ex-ante sample (period 2010–2014), even though most indices in the Ex-post sample are slightly higher. Since the temporal comparisons of genetic diversity of this study are clearly descriptive and depend upon differences in the sample size as well as demographic variation (Landguth et al., 2010; Tallmon et al., 2010), slight temporal differences may be attributed to stochastic effects rather than real genetic changes in the sampled population (Epps & Keyghobadi, 2015). Therefore, P. magdalenae has conserved high genetic diversity in the middle and lower parts of the Cauca River, with no obvious changes detected over the short-term.
Despite the high genetic diversity, this species exhibited high heterozygosity deficit in the Ex-post, the same as in the Ex-ante sample from the Cauca River (Landínez-García et al., 2020) and populations from the Magdalena, Atrato, and Sinú rivers (Fontalvo et al., 2018; Landínez-García et al., 2020; Orozco-Berdugo & Narváez-Barandica, 2014). Possible causes of such deficit include the presence of null alleles, Wahlund effect, and endogamy (Waples, 2015). In contrast, factors less likely to contribute involve assortative mating, as P. magdalenae exhibits external fertilization and follows a seasonal reproductive strategy that involves group spawning in open areas without parental care (Jiménez-Segura et al., 2010; López-Casas et al., 2016), suggesting a high likelihood of random fertilization among the co-migrating individuals.
The presence of null alleles is more likely when using amplification of heterologous markers compared to amplification of species-specific markers (Bhargava & Fuentes, 2010; Guichoux et al., 2011). In the case of P. magdalenae, both species-specific and heterologous markers have been assessed. Specifically, Landínez-García et al. (2020) and the present study found a lower degree of heterozygote deficit when using species-specific markers compared to heterologous microsatellites designed for P. lineatus (Fontalvo et al., 2018; Orozco-Berdugo & Narváez-Barandica, 2014). However, null alleles alone do not explain the significant deficits as they have been observed with both markers. Additionally, some authors have reported minor impacts on genetic differentiation and parentage analyses when using loci with frequencies of null alleles lower than 20% (Chapuis & Estoup, 2007; Huang et al., 2016; Rico et al., 2017).
Alternatively, the heterozygosity deficit may be explained by Wahlund effect due to the coexistence of two genetically distinct populations of P. magdalenae in the Cauca River evidenced by some results of genetic structure. However, the persistence of this deficit when analyzing each group separately indicates that inbreeding may explain the high heterozygosity deficit found in this species, as previously reported for this species (Orozco-Berdugo & Narváez-Barandica, 2014; Fontalvo et al., 2018; Landínez-García et al., 2020) and for such other fishes of the Cauca River as Pimelodus yuma Villa-Navarro & Acero P. 2017 (Joya et al., 2021) and Pimelodus grosskopfii Steindachner 1879 (Restrepo-Escobar et al., 2021). The stocking of highly related individuals is a non-excluding explanation, considering that this species has been a target of restocking efforts in this basin (De La Rosa et al., 2020; Landínez-García et al., 2020; Márquez et al., 2020). Such a high degree of inbreeding is of concern since, according to Franklin (1980) and Soulé (1980), inbreeding values higher than 10% in wild populations may result in adverse fitness and evolutionary effects on the species.
Considering the generalized concept of neutral evolution of microsatellites markers (Oliveira et al., 2006; Putman & Carbone, 2014), the detection of five candidate loci under natural selection for P. magdalenae in the floodplain Grande suggests either hitchhiking or background selection (Cutter & Payseur, 2013; Stephan, 2010). Landínez-García et al. (2020) argued that unidirectional gene flow from hatcheries to the wild populations, among other potential anthropogenic factors, could explain the selection signals and the unexpected genetic structure found in P. magdalenae in the Magdalena River. Candidate loci under natural selection have also been detected in the characid fish Curimata mivartii Steindachner 1878 in Las Culebras, a large floodplain close to Grande, which could be explained by isolation of this site with the Cauca River due to droughts produced by recent ENSO phenomena (Landínez-García & Marquez, 2018). Both unidirectional gene flow and temporal disconnections between floodplain and river could possibly explain the putative selection signals found in the Ex-post sample of P. magdalenae of the Cauca River. Although floodplains are essential habitats within the life cycle of fishes of the Magdalena-Cauca basin (Jiménez-Segura et al., 2020), their ecology and human impacts are largely unexplored, limiting the interpretation of these findings.
Demographic estimates showed that the effective population size (Ne) in the Ex-post sample substantially increased in the short-term using the LD method. Given that Ne is often positively correlated with population size, this result may seem contradictory considering the decreasing annual catches of P. magdalenae reported over the last 40 years in the Magdalena basin (Mojica et al., 2012; Barreto, 2017) and the genetic evidence of recent bottlenecks (Landínez-García et al., 2020; this study). Although the demographic recovery of this population is likely, an alternative explanation also includes the overestimation of Ne. Since the sampled area belongs to a wider geographic range of the Magdalena-Cauca River basin where this species occurs, the studied population is not a closed system, so Ne can be upwardly biased due to a high and constant rate of migration according to Waples & England (2011), who showed that under a total pool of parents larger than the local size, the estimation can converge on a value that represents the metapopulation effective size. Additionally, the coexistence of two mildly differentiated stocks may also explain such overestimation of the overall Ne, as its estimation assumes absence of genetic structure (Waples & Do, 2010; Waples & England, 2011). However, beyond the likely biased estimation, a higher Ne using the LD method is expected to reflect a lower inbreeding rate in the wild population (Ryman et al., 2019). Therefore, the large Ne estimated herein would be concordant with the stable inbreeding indices found in the Cauca River over the short term (Martinez et al., 2018).
In contrast, the relatively small Ne estimate obtained using the temporal method may be downwardly biased because of unexpectedly large variance of the allele frequency changes. Likely biasing factors for the temporal method include high gene flow (Ryman et al., 2014), age-structured population (overlapping generations; Waples & Yokota, 2007), and fluctuating population size or non-severe bottleneck, this latter causing allele frequency changes by drift before some alleles are lost (Freeland, 2020). Given the relevance of the Ne for assessing the evolutionary risks of the species in the conservation context, the lower confidence intervals are a useful and more prudent metric to consider in this case (Waples & Do, 2010). Therefore, the lower confidence value estimated in the temporal method close to 500 may suggest long-term risks for maintenance of the evolutionary potential of P. magdalenae in the Cauca River (Frankham et al., 2014).
Results of the genetic structure analysis suggested high gene flow throughout the studied length of the Cauca River and the absence of spatial genetic structure even between sections upstream and downstream of the dam. This outcome is partly unexpected, as fragmented populations are subjected to some degree of isolation (Liermann et al., 2012; Epps & Keyghobadi, 2015; Barbarossa et al., 2020), causing differences in allele frequencies between fragmented sections due to genetic drift and bottlenecks, especially upstream of dams (Coleman et al., 2018; Vega-Retter et al., 2020; Yamamoto et al., 2004). Similar findings for gene flow in populations of migratory fishes fragmented by dams have been attributed to factors such as fishways or restocking between fragmented sections, permanence of barrier-free tributaries as alternative routes for migration and short-time periods elapsed since the habitat disturbance (Ribolli et al., 2012; Cheng et al., 2013; Ferreira et al., 2017; Prado et al., 2018; Pimentel et al., 2020). In this case, since the Ituango Dam has no fishways, such structures cannot explain the current gene flow of P. magdalenae. However, the recent occurrence of a disturbance on the population might require a number of generations to manifest (a time lag), which limits the likelihood of detecting genetic changes over the short term (Epps & Keyghobadi, 2015). Additionally, this species has a preexistent high gene flow (Landínez-García et al., 2020) and a wide distribution due to the absence of other geographical barriers in most reaches of the Magdalena basin (López-Casas et al., 2016; Zapata & Usma, 2013), factors that can contribute to the maintenance of its genetic variability (Díez-del-Molino et al., 2013; Epps & Keyghobadi, 2015; Thomaz et al., 2016). Furthermore, dispersal or movement through the dam by translocation can also support gene flow. For instance, the detection of seven individuals collected downstream as likely migrants from upstream sections according to the Bayesian analysis of Geneclass2 (Piry et al., 2004) may indicate movement of individuals through the spillway or discharge tunnel of the dam, sampled individuals that migrated before fragmentation (considering a life cycle > 6 years for P. magdalenae in the Cauca River; unpublished data) and drift of larvae from upstream sections, although this last explanation seems unlikely due to the reservoir´s lentic environment (Agostinho et al., 2008). On the other hand, the assignment of one individual collected upstream to downstream sections as its most likely origin place suggests restocking or fish translocation, since such practices might be common in the Magdalena basin (Márquez et al., 2020).
Despite the absence of spatial genetic structure, the coexistence of two genetic stocks of P. magdalenae, an outcome common to both temporal samples, may respond to isolation over time (Hendry & Day, 2005) due to the migratory behavior of this species, wherein distinct groups of individuals may migrate during each of the two flood periods of the Cauca River (Jiménez-Segura et al., 2010, 2016; López-Casas et al., 2016). This pattern was also detected in the congener P. lineatus, showing three genetic stocks that migrate within the lower basin of the Uruguay River according to three seasonal periods (Rueda et al., 2013). Likewise, two stocks were detected in P. costatus within the same reproductive season with high gene flow (Braga-Silva & Galetti, 2016). However, the samples evaluated in this study constitute individuals collected in different years and periods of drought and flood, so this hypothesis must be assessed in further genetic studies that include sampling designs based on the migratory behavior of P. magdalenae. Further, although both stocks showed genetic structure differences over time (as indicated by AMOVA and pairwise genetic indices between temporal samples), no genetic differences were found between fragmented sections (Table 5). Therefore, considering the uncertain biological reasons behind these genetic stocks and the potential impacts of restocking on the analysis of causality performed, this study did not find any apparent short-term effects of population fragmentation.
In conclusion, despite its disrupted migratory route in the Cauca River, P. magdalenae retained high genetic diversity in temporal samples, with stable inbreeding values and high gene flow both upstream and downstream of the Ituango Dam. Likely explanations for these outcomes include large effective population size and insufficient time elapsed since fragmentation to give rise to potential changes. Moreover, the slight variations in the frequencies of the stocks, which may result from restocking effects, are not attributable to the dam or any other habitat disturbances, and require further studies to determine the origin and dynamics of the genetic stocks that coexist in the river.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Change history
29 November 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10750-023-05429-7
References
Agostinho, A. A., F. M. Pelicice & L. C. Gomes, 2008. Dams and the fish fauna of the Neotropical region: Impacts and management related to diversity and fisheries. Brazilian Journal of Biology 68: 1119–1132.
Arthington, A. H., N. K. Dulvy, W. Gladstone & I. J. Winfield, 2016. Fish conservation in freshwater and marine realms: status, threats and management. Aquatic Conservation: Marine and Freshwater Ecosystems 26: 838–857.
Baggio, R. A., S. B. L. Araujo, D. Ayllón & W. A. Boeger, 2018. Dams cause genetic homogenization in populations of fish that present homing behavior: evidence from a demogenetic individual-based model. Ecological Modelling 384: 209–220.
Barbarossa, V., R. J. P. Schmitt, M. A. J. Huijbregts, C. Zarfl, H. King & A. M. Schipper, 2020. Impacts of current and future large dams on the geographic range connectivity of freshwater fish worldwide. Proceedings of the National Academy of Sciences of the United States of America 117: 3648–3655.
Barletta, M., V. E. Cussac, A. A. Agostinho, C. Baigún, E. K. Okada, A. C. Catella, N. F. Fontoura, P. S. Pompeu, L. F. Jiménez-Segura, V. S. Batista, C. A. Lasso, D. Taphorn, & N. N. Fabré, 2015. Fisheries ecology in South American river basins. In Craig, J. F. (ed), Freshwater Fisheries Ecology 76: 311–348.
Barreto, C., 2017. Producción pesquera de la cuenca del río Magdalena: desembarcos y estimación ecosistémica. Informe Técnico Final. The Nature Conservancy, MacArthur Foundation, AUNAP. Bogotá, http://sepec.aunap.gov.co/Home/VerPdf/63.
Beneteau, C. L., N. E. Mandrak & D. D. Heath, 2008. The effects of river barriers and range expansion of the population genetic structure and stability in Greenside Darter (Etheostoma blennioides) populations. Conservation Genetics 10: 477–487.
Bhargava, A. & F. F. Fuentes, 2010. Mutational dynamics of microsatellites. Molecular Biotechnology 40: 250–266.
Braga-Silva, A. & P. M. Galetti, 2016. Evidence of isolation by time in freshwater migratory fish Prochilodus costatus (Characiformes, Prochilodontidae). Hydrobiologia 765: 159–167.
Carvalho-Costa, L. F., T. Hatanaka & P. M. Galetti, 2008. Evidence of lack of population substructuring in the Brazilian freshwater fish Prochilodus costatus. Genetics and Molecular Biology 31: 377–380.
Chapuis, M. P. & A. Estoup, 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution Oxford Academic 24: 621–631.
Cheng, F., W. Li, Q. Wu, E. Hallerman & S. Xie, 2013. Microsatellite DNA variation among samples of bronze gudgeon, Coreius heterodon, in the mainstem of the Yangtze River, China. Ichthyological Research 60: 165–171.
Coimbra, M. R. M., A. P. S. Lima, K. K. C. Oliveira & W. Severi, 2017. Microsatellite assessment of the genetic diversity in indigenous populations of curimba (Prochilodus argenteus) in the São Francisco river (Brazil). Conservation Genetics 18: 965–975.
Coleman, R. A., B. Gauffre, A. Pavlova, L. B. Beheregaray, J. Kearns, J. Lyon, M. Sasaki, R. Leblois, C. Sgro & P. Sunnucks, 2018. Artificial barriers prevent genetic recovery of small isolated populations of a low-mobility freshwater fish. Heredity 120: 515–532.
Cornuet, J. M. & G. Luikart, 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Cutter, A. D. & B. A. Payseur, 2013. Genomic signatures of selection at linked sites: Unifying the disparity among species. Nature Reviews Genetics 14: 262–274.
De La Rosa, J., P. P. Fontalvo & G. Orozco-Berdugo, 2020. Caracterización genética de reproductores de Prochilodus magdalenae (Pisces: Prochilodontidae) usados en programas de repoblamiento en Colombia. Journal of Basic and Applied Genetics 31: 53–63.
Deinet, S., K. Scott-Gatty, H. Rotton, W. M. Twardek, V. Marconi, L. McRae, L. J. Baumgartner, K. Brink, J. E. Claussen, S. J. Cooke, W. Darwall, B. K. Eriksson, C. Garcia de Leaniz, Z. Hogan, J. Royte, L. G. M. Silva, M. L. Thieme, D. Tickner, J. Waldman, H. Wanningen, O. L. F. Weyl, & A. Berkhuysen, 2020. The Living Planet Index (LPI) for migratory freshwater fish - Technical Report. World Fish Migration Foundation. pp. 7–35. https://worldfishmigrationfoundation.com/wp-content/uploads/2020/07/LPI_report_2020.pdf
Díez-del-Molino, D., G. Carmona-Catot, R.-M. Araguas, O. Vidal, N. Sanz, E. García-Berthou & J.-L. García-Marín, 2013. Gene flow and maintenance of genetic diversity in invasive mosquitofish (Gambusia holbrooki). PLoS ONE 8: e82501.
Do, C., R. S. Waples, D. Peel, G. M. Macbeth, B. J. Tillett & J. R. Ovenden, 2014. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14: 209–214.
Duarte, L. O., L. Manjarrés–Martínez, & H. Reyes-Ardila, 2019. Estadísticas de desembarco y esfuerzo de las pesquerías artesanales e industriales de Colombia entre febrero y diciembre de 2019. Servicio Estadístico Pesquero Colombiano SEPEC. Bogotá D.C., Colombia.
Dudgeon, D., A. H. Arthington, M. O. Gessner, Z. I. Kawabata, D. J. Knowler, C. Lévêque, R. J. Naiman, A. H. Prieur-Richard, D. Soto, M. L. J. Stiassny & C. A. Sullivan, 2006. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biological Reviews of the Cambridge Philosophical Society 81: 163–182.
Epps, C. W. & N. Keyghobadi, 2015. Landscape genetics in a changing world: disentangling historical and contemporary influences and inferring change. Molecular Ecology 24: 6021–6040.
Evanno, G., S. Regnaut & J. Goudet, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620.
Excoffier, L. & H. E. L. Lischer, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
Ferreira, D. G., L. Souza-Shibatta, O. A. Shibatta, S. H. Sofia, J. Carlsson, J. H. P. Dias, S. Makrakis & M. C. Makrakis, 2017. Genetic structure and diversity of migratory freshwater fish in a fragmented Neotropical river system. Reviews in Fish Biology and Fisheries 27: 209–231.
Fluker, B. L., B. R. Kuhajda & P. M. Harris, 2014. The effects of riverine impoundment on genetic structure and gene flow in two stream fishes in the Mobile River basin. Freshwater Biology 59: 526–543.
Foll, M. & O. Gaggiotti, 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993.
Fontalvo, P. P., G. Orozco-Berdugo & J. Narváez-Barandica, 2018. Diversidad y estructura genética del Prochilodus magdalenae (Pisces: Prochilodontidae) aguas arriba y abajo de la represa Betania, Colombia. Intropica 13: 87–100.
Frankham, R., C. J. A. Bradshaw & B. W. Brook, 2014. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biological Conservation 170: 56–63.
Franklin, I. R., 1980. Evolutionary change in small populations. In Soulé, M. E. & B. A. Wilcox (eds), Conservation Biology: An Evolutionary-Ecological Perspective Sinauer Associates, Sunderland: 135–149.
Freeland, J. R., 2020. Genetic analysis of single populations. In Freeland, J. R. (ed), Molecular Ecology Wiley, New York: 149–189.
Galvis, G. & J. I. Mojica, 2007. The Magdalena River fresh water fishes and fisheries. Aquatic Ecosystem Health & Management 10: 127–139.
García-Alzate, C., C. DoNascimiento, F. A. Villa-Navarro, J. E. García-Melo, & G. Herrera R., 2020. Diversidad de peces de la cuenca de Río Magdalena, Colombia. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. Bogotá, D.C. pp. 85–113.
Garza, J. C. & E. G. Williamson, 2001. Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318.
Goudet, J., 2003. Fstat (ver. 2.9.4), a program to estimate and test population genetics parameters. https://www2.unil.ch/popgen/softwares/fstat.htm.
Grill, G., B. Lehner, A. E. Lumsdon, G. K. Macdonald, C. Zarfl & C. Reidy Liermann, 2015. An index-based framework for assessing patterns and trends in river fragmentation and flow regulation by global dams at multiple scales. Environmental Research Letters 10: 015001.
Grill, G., B. Lehner, M. Thieme, B. Geenen, D. Tickner, F. Antonelli, S. Babu, P. Borrelli, L. Cheng, H. Crochetiere, H. Ehalt Macedo, R. Filgueiras, M. Goichot, J. Higgins, Z. Hogan, B. Lip, M. E. McClain, J. Meng, M. Mulligan, C. Nilsson, J. D. Olden, J. J. Opperman, P. Petry, C. Reidy Liermann, L. Sáenz, S. Salinas-Rodríguez, P. Schelle, R. J. P. Schmitt, J. Snider, F. Tan, K. Tockner, P. H. Valdujo, A. van Soesbergen & C. Zarfl, 2019. Mapping the world’s free-flowing rivers. Nature 569: 215–221.
Guichoux, E., L. Lagache, S. Wagner, P. Chaumeil, P. Léger, O. Lepais, C. Lepoittevin, T. Malausa, E. Revardel, F. Salin & R. J. Petit, 2011. Current trends in microsatellite genotyping. Molecular Ecology Resources 11: 591–611.
Hendry, A. P. & T. Day, 2005. Population structure attributable to reproductive time: Isolation by time and adaptation by time. Molecular Ecology 14: 901–916.
Herrera-Pérez, J., J. L. Parra, D. Restrepo-Santamaría & L. F. Jiménez-Segura, 2019. The influence of abiotic environment and connectivity on the distribution of diversity in an Andean fish fluvial network. Frontiers in Environmental Science 7: 1–9.
Hilsdorf, A. W. S. & E. M. Hallerman, 2017. Characterization of Genetic Resources. In Hilsdorf, A. W. S. & E. M. Hallerman (eds), Genetic Resources of Neotropical Fishes Springer International Publishing, Switzerland: 55–117.
Huang, K., K. Ritland, D. W. Dunn, X. Qi, S. Guo & B. Li, 2016. Estimating relatedness in the presence of null alleles. Genetics 202: 247–260.
IUCN Standards and Petitions Committee., (2019). Guidelines for Using the IUCN Red List Categories and Criteria. Version 14. http://www.iucnredlist.org/documents/RedListGuidelines.pdf.
Iwersen, L. L. H., C. M. R. De Melo, C. Lazoski, E. Zaniboni-Filho & J. Ribolli, 2019. Genetic implications of restocking programs on wild populations of streaked prochilod Prochilodus lineatus. Boletim Do Instituto De Pesca 45: 1–14.
Jiménez-Segura, L. F., G. Galvis-Vergara, P. Cala-Cala, C. A. García-Alzate, S. López-Casas, M. I. Ríos-Pulgarín, G. A. Arango, N. J. Mancera-Rodríguez, F. Gutiérrez-Bonilla & R. Álvarez-León, 2016. Freshwater fish faunas, habitats and conservation challenges in the Caribbean river basins of north-western South America. Journal of Fish Biology 89: 65–101.
Jiménez-Segura, L. F., J. Palacio & R. Leite, 2010. River flooding and reproduction of migratory fish species in the Magdalena River basin, Colombia. Ecology of Freshwater Fish 19: 178–186.
Jiménez-Segura, L., J. Herrera-Pérez, D. Valencia-Rodríguez, I. Castaño-Tenorio, S. López-Casas, M. I. Ríos, Y. F. Rondón-Martínez, K. Rivera-Coley, J. Morales, M. Arboleda, S. Muñoz-Duque, V. Atencio, A. F. Galeano-Moreno, R. Valbuena, J. Escobar, J. Ospina-Pabón, L. García-Melo, D. Gualtero, J. C. Alonso, & D. Restrepo- Santamaría, 2020. Ecología e historias de vida de los peces en la cuenca del río Magdalena, Colombia. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río
Jombart, T., 2008. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405.
Jorde, P. E. & N. Ryman, 2007. Unbiased estimator for genetic drift and effective population size. Genetics 177: 927–935.
Jost, L., 2008. GST and its relatives do not measure differentiation. Molecular Ecology 17: 4015–4026.
Joya, C. D., R. M. Landínez-García & E. J. Márquez, 2021. Development of microsatellite loci and population genetics of the catfish Pimelodus yuma (Siluriformes: Pimelodidae). Neotropical Ichthyology 19: e200114.
Klütsch, C. F. C., S. N. Maduna, N. Polikarpova, K. Forfang, P. E. Aspholm, T. Nyman, H. G. Eiken, P. Amundsen & S. B. Hagen, 2019. Genetic changes caused by restocking and hydroelectric dams in demographically bottlenecked brown trout in a transnational subarctic riverine system. Ecology and Evolution 9: 6068–6081.
Laikre, L., M. K. Schwartz, R. S. Waples & N. Ryman, 2010. Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends in Ecology and Evolution 25: 520–529.
Landguth, E. L., S. A. Cushman, M. K. Schwartz, K. S. Mckelvey, M. Murphy & G. Luikart, 2010. Quantifying the lag time to detect barriers in landscape genetics. Molecular Ecology 19: 4179–4191.
Landínez-García, R. M., & E. J. Márquez, 2016. Development and characterization of 24 polymorphic microsatellite loci for the freshwater fish Ichthyoelephas longirostris (Characiformes: Prochilodontidae). PeerJ 4: e2419. https://peerj.com/articles/2419.
Landínez-García, R. M. & E. J. Marquez, 2018. Microsatellite loci development and population genetics in Neotropical fish Curimata mivartii (Characiformes: Curimatidae). PeerJ 6: e5959.
Landínez-García, R. M., J. C. Narváez & E. J. Márquez, 2020. Population genetics of the freshwater fish Prochilodus magdalenae (Characiformes: Prochilodontidae), using species-specific microsatellite loci. PeerJ 8: e10327.
Lasso, C. A., E. Agudelo Córdoba, L. F. Jiménez-Segura, H. Ramírez-Gil, M. Morales-Betancourt, R. E. Ajiaco-Martínez, F. de Paula Gutiérrez, J. S. Usma Oviedo, S. E. Muñoz Torres, & A. I. Sanabria Ochoa, 2011. I. Catálogo de los recursos pesqueros continentales de Colombia. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. I Serie Editorial Recursos Hidrobiológicos y pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá DC: p. 715.
Li, Y. L. & J. X. Liu, 2018. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Molecular Ecology Resources 18: 176–177.
Liermann, C. R., C. Nilsson, J. Robertson & R. Y. Ng, 2012. Implications of dam obstruction for global freshwater fish diversity. BioScience 62: 539–548.
Liu, D., X. Li & Z. Song, 2020. No decline of genetic diversity in elongate loach (Leptobotia elongata) with a tendency to form population structure in the upper Yangtze River. Global Ecology and Conservation 23: e01072.
Lopera-Barrero, N. M., F. P. de Souza, E. C. S. de Lima, A. M. Urrea-Rojas, P. L. de Castro, E. S. dos Reis Goes, V. C. F. Pandolfi, A. L. Yamachita, C. A. L. de Oliveira, N. G. Leite & R. P. Ribeiro, 2019. Genetic Diversity of Migratory Fish Populations of the Rio Grande Reservoir (São Paulo, Brazil), Ciências Agrárias, Semina:, 503–510.
Lopera-Barrero, N. M., S. C. A. Santos, E. S. R. Goes, P. L. Castro, F. P. Souza, A. R. Poveda-Parra, J. Casseta, B. G. Pontillo & R. P. Ribeiro, 2016. Monitoramento e conservação genética de populações naturais de Prochilodus lineatus dos rios Pardo, Mogi-Guaçu e Tietê, São Paulo. Arquivo Brasileiro De Medicina Veterinária e Zootecnia 68: 1621–1628.
López-Casas, S., L. F. Jiménez-Segura, A. A. Agostinho & C. M. Pérez, 2016. Potamodromous migrations in the Magdalena River basin: Bimodal reproductive patterns in Neotropical rivers. Journal of Fish Biology 89: 157–171.
López-Casas, S., Y. F. Rondón-Martínez, A. Gutiérrez-Cortés, J. L. Escobar-Cardona, S. Muñoz-Duque, D. Valencia-Rodríguez, P. Petry, A. M. Batista-Morales, C. Rincón, L. F. Casas, J. G. Ospina-Pabón, V. Atencio-García, M. Valderrama Barco, C. A. Lasso, & L. F. Jiménez-Segura, 2020. Diagnóstico del grado de amenaza y medidas de manejo para los peces del río Magdalena, Colombia. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, DC.: pp. 391–429.
Luikart, G. & J. M. Cornuet, 1998. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology 12: 228–237.
Manel, S., M. K. Schwartz, G. Luikart & P. Taberlet, 2003. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology and Evolution 18: 189–197.
Márquez, E. J., N. Restrepo-Escobar, A. J. Yepes-Acevedo, & J. C. Narváez, 2020. Diversidad y estructura genética de los peces de la cuenca del Magdalena, Colombia. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, DC: pp. 115–157.
Martinez, A. S., J. R. Willoughby & M. R. Christie, 2018. Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecology and Evolution 8: 12022–12031.
Meirmans, P. G., 2006. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60: 2399–2402.
Meirmans, P. G. & P. W. Hedrick, 2011. Assessing population structure: FST and related measures. Molecular Ecology Resources 11: 5–18.
Melo, B. F., Y. Sato, F. Foresti & C. Oliveira, 2013. The roles of marginal lagoons in the maintenance of genetic diversity in the Brazilian migratory fishes Prochilodus argenteus and P Costatus. Neotropical Ichthyology 11: 625–636.
Mojica, J. I., J. S. Usma, R. Álvarez-León, & C. A. Lasso, 2012. Libro rojo de peces dulceacuícolas de Colombia 2012. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, D.C
Oliveira, E. J., J. G. Pádua, M. I. Zucchi, R. Vencovsky & M. L. C. Vieira, 2006. Origin, evolution and genome distribution of microsatellites. Genetics and Molecular Biology 29: 294–307.
Orozco-Berdugo, G. & J. C. Narváez-Barandica, 2014. Genetic diversity and population structure of bocachico Prochilodus magdalenae (Pisces, Prochilodontidae) in the Magdalena river basin and its tributaries, Colombia. Genetics and Molecular Biology 37: 37–45.
Paetkau, D., R. Slade, M. Burden & A. Estoup, 2004. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Molecular Ecology 13: 55–65.
Pavlova, A., L. B. Beheregaray, R. Coleman, D. Gilligan, K. A. Harrisson, B. A. Ingram, J. Kearns, A. M. Lamb, M. Lintermans, J. Lyon, T. T. T. Nguyen, M. Sasaki, Z. Tonkin, J. D. L. Yen & P. Sunnucks, 2017. Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: a call for assisted gene flow. Evolutionary Applications 10: 531–550.
Peakall, R. & P. E. Smouse, 2006. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295.
Peakall, R. & P. E. Smouse, 2012. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539.
da Pimentel, J. & S. M., S. Ludwig, L. C. Resende, P. F. P. Brandão-Dias, A. H. Pereira, N. L. de Abreu, I. C. Rosse, A. P. V. Martins, S. Facchin, J. de M. Lopes, G. B. Santos, C. B. M. Alves, & E. Kalapothakis, 2020. Genetic evaluation of migratory fish: Implications for conservation and stocking programs. Ecology and Evolution 10: 10314–10324.
Piry, S., A. Alapetite, J. M. Cornuet, D. Paetkau, L. Baudouin & A. Estoup, 2004. GENECLASS2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity 95: 536–539.
Piry, S., G. Luikart & J. M. Cornuet, 1999. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity 90: 502–503.
Prado, F. D., R. Fernandez-Cebrián, F. Foresti, C. Oliveira, P. Martínez & F. Porto-Foresti, 2018. Genetic structure and evidence of anthropogenic effects on wild populations of two Neotropical catfishes: baselines for conservation. Journal of Fish Biology 92: 55–72.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Puechmaille, S. J., 2016. The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Molecular Ecology Resources 16: 608–627.
Putman, A. I. & I. Carbone, 2014. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecology and Evolution 4: 4399–4428.
Raymond, M. & F. Rousset, 1995. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249.
Reid, A. J., A. K. Carlson, I. F. Creed, E. J. Eliason, P. A. Gell, P. T. J. Johnson, K. A. Kidd, T. J. MacCormack, J. D. Olden, S. J. Ormerod, J. P. Smol, W. W. Taylor, K. Tockner, J. C. Vermaire, D. Dudgeon & S. J. Cooke, 2019. Emerging threats and persistent conservation challenges for freshwater biodiversity. Biological Reviews 94: 849–873.
Reis, R. E., J. S. Albert, F. Di Dario, M. M. Mincarone, P. Petry & L. A. Rocha, 2016. Fish biodiversity and conservation in South America. Journal of Fish Biology 89: 12–47.
Restrepo, J. D., A. Cárdenas-Rozo, J. F. Paniagua-Arroyave, & L. Jiménez-Segura, 2020. Aspectos físicos de la cuenca del río Magdalena, Colombia: geología, hidrología, sedimentos, conectividad, ecosistemas acuáticos e implicaciones para la biota. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, DC: pp. 41–83.
Restrepo-Escobar, N., A. J. Yepes-Acevedo & E. J. Márquez, 2021. Population genetics of three threatened catfish species in heterogeneous environments of the Cauca River Colombia. Neotropical Ichthyology 19: 2021.
Ribolli, J., C. M. R. de Melo & E. Zaniboni-Filho, 2012. Genetic characterization of the Neotropical catfish Pimelodus maculatus (Pimelodidae, Siluriformes) in the Upper Uruguay River. Genetics and Molecular Biology 35: 761–769.
Rico, C., J. A. Cuesta, P. Drake, E. Macpherson, L. Bernatchez, & A. D. Marie, 2017. Null alleles are ubiquitous at microsatellite loci in the Wedge Clam (Donax trunculus). PeerJ: e3188.
Rodríguez-Olarte, D., J. I. Mojica & D. Taphorn, 2011. Northern South America Magdalena and Maracaibo Basins. In Albert, J. S. & R. E. Reis (eds), Historical Biogeography of Neotropical Freshwater Fishes University of California Press, Los Angeles: 243–257.
Rousset, F., 2008. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8: 103–106.
Rueda, E. C., P. Carriquiriborde, A. M. Monzón, G. M. Somoza & G. Ortí, 2013. Seasonal variation in genetic population structure of sábalo (Prochilodus lineatus) in the Lower Uruguay River. Genética 141: 401–407.
Ruzich, J., K. Turnquist, N. Nye, D. Rowe & W. A. Larson, 2019. Isolation by a hydroelectric dam induces minimal impacts on genetic diversity and population structure in six fish species. Conservation Genetics 20: 1421–1436.
Ryman, N., F. W. Allendorf, P. E. Jorde, L. Laikre & O. Hössjer, 2014. Samples from subdivided populations yield biased estimates of effective size that overestimate the rate of loss of genetic variation. Molecular Ecology Resources 14: 87–99.
Ryman, N., L. Laikre & O. Hössjer, 2019. Do estimates of contemporary effective population size tell us what we want to know? Molecular Ecology 28: 1904–1918.
Sanches, A., P. M. Galetti, F. Galzerani, J. Derazo, B. Cutilak-Bianchi & T. Hatanaka, 2012. Genetic population structure of two migratory freshwater fish species (Brycon orthotaenia and Prochilodus argenteus) from the São Francisco River in Brazil and its significance for conservation. Latin American Journal of Aquatic Research 40: 177–186.
Schwartz, M. K., G. Luikart & R. S. Waples, 2007. Genetic monitoring as a promising tool for conservation and management. Trends in Ecology and Evolution 22: 25–33.
Soulé, M. E., 1980. Thresholds for survival: Maintaining fitness and evolutionary potential. In Soulé, M. E. & B. A. Wilcox (eds), Conservation Biology: An Evolutionary Ecological Perspective Sinauer Associates, Sunderland: 151–169.
Stephan, W., 2010. Genetic hitchhiking versus background selection: the controversy and its implications. Philosophical Transactions of the Royal Society B: Biological Sciences 365: 1245–1253.
Tallmon, D. A., D. Gregovich, R. S. Waples, C. Scott Baker, J. Jackson, B. L. Taylor, E. Archer, K. K. Martien, F. W. Allendorf & M. K. Schwartz, 2010. When are genetic methods useful for estimating contemporary abundance and detecting population trends? Molecular Ecology Resources 10: 684–692.
Tamario, C., J. Sunde, E. Petersson, P. Tibblin & A. Forsman, 2019. Ecological and evolutionary consequences of environmental change and management actions for migrating fish. Frontiers in Ecology and Evolution 7: 1–27.
Thomaz, A. T., M. R. Christie & L. L. Knowles, 2016. The architecture of river networks can drive the evolutionary dynamics of aquatic populations. Evolution 70: 731–739.
Valderrama, M., J. L. Escobar C., R. Pardo B., M. Toro S., J. C. Gutiérrez C., & S. López C., 2020. Servicios ecosistémicos generados por los peces de la cuenca del río Magdalena, Colombia. In Jiménez-Segura, L., & C. A. Lasso (eds), XIX. Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, DC: pp. 205–235.
Van Leeuwen, C. H. A., K. Dalen, J. Museth, C. Junge & L. A. Vøllestad, 2018. Habitat fragmentation has interactive effects on the population genetic diversity and individual behaviour of a freshwater salmonid fish. River Research and Applications 34: 60–68.
Van Oosterhout, C., W. F. Hutchinson, D. P. M. Wills & P. Shipley, 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538.
Vega-Retter, C., P. Muñoz-Rojas, N. Rojas-Hernández, S. Copaja, L. Flores-Prado & D. Véliz, 2020. Dammed river: Short- and long-term consequences for fish species inhabiting a river in a Mediterranean climate in central Chile. Aquatic Conservation: Marine and Freshwater Ecosystems 30: 2254–2268.
Waples, R. S., 2015. Testing for Hardy-Weinberg proportions: Have we lost the plot? Journal of Heredity 106: 1–19.
Waples, R. S. & C. Do, 2010. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evolutionary Applications 3: 244–262.
Waples, R. S. & P. R. England, 2011. Estimating contemporary effective population size on the basis of linkage disequilibrium in the face of migration. Genetics 189: 633–644.
Waples, R. S. & M. Yokota, 2007. Temporal estimates of effective population size in species with overlapping generations. Genetics 175: 219–233.
WWF, 2020. Living Planet Report 2020 - Bending the curve of biodiversity loss. WWF, Gland, https://www.zsl.org/sites/default/files/LPR2020Fullreport.pdf.
Yamamoto, S., K. Morita, I. Koizumi & K. Maekawa, 2004. Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: Spatial-temporal changes in gene frequencies. Conservation Genetics 5: 529–538.
Yamamoto, S., K. Morita, & G. Sahashi, 2019. Spatial and temporal changes in genetic structure and diversity of isolated white-spotted charr (Salvelinus leucomaenis) populations. Hydrobiologia 840: 35–48. https://link-springer-com.ezproxy.unal.edu.co/article/10.1007/s10750-019-3924-9.
Zapata, L. A., & J. S. Usma (eds), 2013. Guía de las especies migratorias de la biodiversidad en Colombia. Peces. Vol. 2. Ministerio de Ambiente y Desarrollo Sostenible/WWF-Colombia. Bogotá DC
Acknowledgements
The authors thank the Universidad de Antioquia, Universidad de Córdoba, Fundación Humedales, Integral S.A.S and Ana María Restrepo and Giovanny Olaya, for providing the samples used in the analyses of this study. The authors also thank Omer Campo Nieto and Ricardo Landínez García for their support in the laboratory work.
Funding
Open Access funding provided by Colombia Consortium. This study was funded by Universidad Nacional de Colombia Sede Medellín and Empresas Públicas de Medellín, Grant CT-2019-000661 “Variabilidad genética de un banco de peces de los sectores medio y bajo del Río Cauca.”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Handling editor: Christian Sturmbauer
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: the hyphen was missing from the first author's family name and the word "characiform" was not removed from the 5th paragraph, line 11 of the Discussion section.
Supplementary Information
Below is the link to the electronic supplementary material.
10750_2023_5396_MOESM1_ESM.docx
Outlier loci in the Grande floodplain collection within section S6 of the Cauca River, Colombia. Outlier loci are shown in bold. PP: Posterior probability of the model including selection. Log (PO): Base-10 logarithm of the ratio between posterior odds of the model with selection and neutrality (PO). q-value: Minimum false discovery rate (FDR) at which the locus may become significant. Alpha: Specific component of each locus indicating strength and direction of selection; positive values suggesting diversifying selection and negative values suggesting purifying or balancing selection. FST: Population-specific component indicating allelic differences between the common gene pool and each collection. Supplementary file1 (DOCX 14 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
García-Castro, K.L., Márquez, E.J. Temporal-scale assessment of population genetics of the freshwater fish Prochilodus magdalenae in an area impacted by construction of a dam. Hydrobiologia 851, 1513–1531 (2024). https://doi.org/10.1007/s10750-023-05396-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05396-z