Abstract
Natural interspecific hybridization between Typha latifolia L. and Typha angustifolia L. was analyzed by morpho-anatomical and molecular methods to determine whether the hybrid Typha glauca Godr. is present in Poland and to identify the best diagnostic traits for its identification. Eighty-three samples of the Typha species were collected. Nine random amplified polymorphic DNA (RAPD) primers provided 12 fragments specific for T. angustifolia and eight fragments specific for T. latifolia. DNA of all sampled individuals was analyzed with 20 diagnostic RAPD markers. The morpho-anatomical variability of T. glauca F1 was found to be quite similar to that observed in parental plants. All of the 41 traits examined in the hybrids overlapped with those observed in the parents, however, the hybrids were visibly closer to T. angustifolia than to T. latifolia. The most discriminate characteristics were the length and pedicel width, as well as the epidermal cell thickness located above vascular bundles in leaf blades. Moreover, preliminary observations of seed sculpture showed that the length of testa cells could also be used to identify T. glauca. Clusters and the hybrid index (for molecular and morphological data) were highly coincident and support the hybridization hypothesis.
Similar content being viewed by others
Introduction
The genus Typha consists of about 30 species (Govaerts, 2012) occurring in almost every part of the world. The majority of cattail species can be found in the wetlands of the temperate northern hemisphere. Among the most common and cosmopolitan species are the broad-leaved cattail (Typha latifolia L.) and the narrow-leaved cattail (Typha angustifolia L.) that occupy extensive areas of the temperate northern hemisphere in Europe and North America. Spontaneous hybrids between both of the species were found in North America in 1844 and in 1889 in Europe, and named Typha glauca Godr. (Smith, 1967). In North America, all three species and, in particular, Typha glauca, are regarded as expansive weeds that have altered wetland ecosystems (Grace & Harrison, 1986; Galatowitsch et al., 1999; Zedler & Kercher, 2004; Angeloni et al., 2006; Shih & Finkelstein, 2008; Tuchman et al., 2009; Lishawa et al., 2010; Travis et al., 2011; Larkin et al., 2012). Reports of spontaneous hybrid occurrence in Europe are comparatively much less frequent. The hybrid is reported to occur in South-Western European locations (Fournier, 1961), in Northern Europe (Luther, 1947), and in Eastern Europe (Kapitonova et al., 2012). Relatively little is known about actual spread of Typha glauca in Central Europe, with only one old report from Germany being available (Figert, 1890). Neither local nor regional studies of flora or the checklists developed for Poland (Mirek et al., 2002) indicate the presence of this species in Poland. However, our field observations carried out in previous years suggest that Typha glauca does grow in this part of Europe. We, therefore, sought for evidence of the existence of Typha glauca as a hybrid even if it remains an unrecognized taxon.
The identification of interspecific hybrids on the basis of morphological and anatomical characteristics may be a challenge. The reason for the difficulties lies in morphological overlapping of the hybrid and the parents (Clausen, 1962), the substantial variability of parent and hybrid characteristics (Carney et al., 2000; González-Rodrίquez et al., 2004), the gradation of intermediate characteristics (Tovar-Sánchez & Oyama, 2004; Stace, 2005), as well as backcrossing (Pacheco et al., 1991). The commonly used diagnostic traits include none that would differentiate between Typha glauca and its parental species (Smith, 1967). The variability of the architectures of Typha ramets and clones is due to a number of factors such as climate and salinity (McNaughton, 1966), as well as population age (Travis et al., 2011). However, the most influential factor impacting on the general habits of the cattails seems to be water depth (Grace & Harrison, 1986; Waters & Shaw, 1990, 1991; Farrell et al., 2010).
The hybrid Typha glauca is common in many regions where the two parental species are sympatric (Galatowitsch et al., 1999; Travis et al., 2010; Kirk et al., 2011), although hybridization is not ubiquitous: in some areas of sympatry, no hybrids have been detected (Selbo & Snow, 2004; Tsyusko et al., 2005), and where they do occur, the frequency of hybrids can vary considerably between regions (Kirk et al., 2011). Some authors claim that the divergence of flowering time may be a critical barrier to species hybridization (Lamont et al., 2003). However, in case of Typha spp., differences in flowering times in Poland are small, and therefore, unlikely to prevent the formation of hybrids. All of the hybrids genotyped by Ball & Freeland (2013) had Typha angustifolia cpDNA sequences. Previous studies have shown that cpDNA is maternally inherited in Typha spp. (Corriveau & Coleman, 1988; Kuehn et al., 1999). Therefore, some form of reproductive barrier prevents either the fertilization of Typha latifolia female flowers by Typha angustifolia pollen, or the subsequent development of viable seeds. Such asymmetrical hybridization has been repeatedly documented in plants (Rieseberg & Carney, 1998) and may result from a number of factors. The latter study concluded that contrary to earlier reports, flowering time is not a barrier to the formation of Typha glauca, (Ball & Freeland, 2013) although an unidentified barrier does prevent the formation of viable hybrids with female Typha angustifolia and male Typha latifolia parents.
The taxonomic status of Typha glauca has long been disputed. Conflicting theories still persist that propose to explain the hybrid nature of Typha glauca, although it is generally accepted to be a form of a hybrid between T. latifolia and T. angustifolia. Mostly Typha glauca has been described as a hybrid species and an F1 hybrid (Bayly & O’Neill, 1971; Lee, 1975; Smith, 1987; Kuehn & White, 1999; Smith, 2000; Snow et al., 2010), or as a species produced by an introgression (Fassett and Calhoun, 1952; Lee, 1975). Moreover, the going assumption until recently was that Typha glauca occurred only as F1 progeny of T. latifolia and T. angustifolia as F1 hybrids were considered to be almost universally sterile (Marsh, 1962; Smith, 1987, 2000). Travis et al. (2010) showed that backcrosses do exist in natural populations. The results of Snow et al. (2010) also confirmed that hybrid populations can include substantial numbers of backcrossed genotypes. Most recently, studies by Kirk et al. (2011) imply that the Typha glauca hybrid is not universally sterile, as suggested by Smith (1987), and that the levels of putative backcrossing found in their study exceed those discovered by Travis et al. (2010) in the western Great Lakes region, which suggests that levels of backcrossing may be non-uniform across different areas of the Great Lakes. Backcrossing can lead to a reshuffling of parental genes; this can, in turn, allow hybrids to express environmental tolerances and growth characteristics that exceed those found in parental species. This may explain the invasive nature of hybrids in the described region. If Typha glauca can produce advanced generation hybrids and/or facilitate the introgression of genes from one parental species to another, their potential to contribute to adaptive evolution and invasiveness within the species complex may be increased (Ellstrand & Schierenbeck, 2000; Rieseberg et al., 2007).
The hypothesis that hybridization occurs in the Polish population of Typha relies on the presence of individuals with intermediate morphological traits in sympatric populations. However, their parentage has not been confirmed using molecular techniques and other diagnostic characteristics. The test conducted for these purposes provides evidence of hybridization and potential introgression. It relies on a combination of nuclear markers and morphological traits. To distinguish between different hypotheses of hybridization and confirm the existence of the hybrid taxon in the examined plant population, it is necessary to identify species-specific (diagnostic) markers for T. latifolia and T. angustifolia. Similarly to Kuehn et al. (1999), we have developed random amplified polymorphic DNA (RAPD) markers specific to the parental species to assess hybrids. We provided a statistical verification of the hitherto applied diagnostic characteristics of Typha angustifolia, Typha latifolia, and Typha glauca and also suggested several new diagnostic characteristics on the basis of microscopic observations of leaves and seed sculpture.
Methods
Data collection
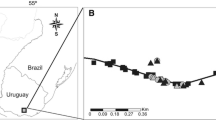
The input data for this study were derived from 83 cattail specimens (Fig. 1, Supplementary material 1). As mentioned in the introduction, we relied on prior field observations in making the assumption that Typha glauca can be found in Midwestern Poland (the Regions of Wielkopolska and Silesia). For this reason, it was in this part of Poland that the majority of the plants were collected. A total of 69 specimens were collected in 20 dispersed sites. Twenty-seven of these came from ponds, 26 specimens were taken from groups of ponds, seven specimens were taken from lakes, five specimens were taken from ditches, and four specimens were taken from rivers.
The plants were collected in August 2010 during the biomass peak. Sprouts were collected of any cattail species identified in each reservoir. In cases where group of reservoirs were found or where a single reservoir was large and harbored a particularly abundant population of cattails, several specimens (ramets) of each species were collected. To avoid collecting ramets of the same clone in such locations, a minimum distance of 30 meters was maintained between the collected ramets. Such a minimum distance was based on a study by Travis et al. (2011). Nevertheless, the actual distances often exceeded 50 m. Specimens were classified according to their morphological structures ranging from broad-leaved to narrow-leaved cattails. Any plants having an intermediate morphological structure of leaves were regarded as hybrids. This sampling scheme was used to collect 69 cattails. Field diagnosis showed that a total sample of 69 cattails consisted of 30 specimens of Typha latifolia, 28 specimens of Typha angustifolia, as well as 11 specimens of their putative hybrid.
In order to check if hybrids can be also find in other regions of Poland random search was run out in the region of Western Poland. We found four reservoirs (ditch, estuary and ponds) in the Pomerania Region which were home to putative hybrids and collected nine specimens of the supposed Typha glauca. We also examined the holdings of the Polish herbarium to find another five putative hybrids at the Jagiellonian University of Cracow.
Measurements
Seventy-eight specimens, including all except the five herbarium samples were measured. The herbarium samples were disregarded as their measurements would be partially incomparable with measurements of fresh plants (e.g., width of female spike in case of flatten spike), would be incomplete (e.g., shoot height in case of cut plant) or difficult to conduct without damaging samples (e.g., plume length, leaf protrusion above the stem).
The measurements of macro-morphological traits relied on a fresh sample and included traits that are commonly used to identify cattail and traits and could be used in the field. We considered the 14 traits of shoot height, shoot diameter, the length and width of the 1st and the 4th leaf, leaf protrusion above the stem, female spike length and width, male spike length, spike interval, the diameter of the inflorescence rachis with pedicels, inflorescence rachis diameter, and plume length. The measurements were carried out by means of tape measure and a slide caliper. Plume length was measured with plotting paper. As plume length varies depending on its location in the inflorescence, each plume specimen was collected at 30 random locations. The average for such 30 values was regarded to be the value of the characteristic. Any qualitative and subjective characteristics such as spandex color were disregarded.
Pedicel measurements, as well as observations of the anatomical structure of leaves relied on a random sample of 20 Typha angustifolia specimens, 20 Typha latifolia specimens, as well as all of F1 specimens of Typha glauca. Observations of leaves anatomy were surveyed on leaves from the middle part of stem. A sliding microtome was used to cut sections to the thickness of 50 μm. Cross-sections of the leaf blade border zone were then displayed. The measurements were carried out using a light microscope with a scale. The measurements covered 21 characteristics: the length and width of margin compartment (MC), compartment 1 (C1), compartment 2 (C2), and compartment 3 (C3); the number of vascular bundles in MC, C1, C2, and C3; the length of the sclerenchyma zone in MC; the width of the beam between MC and C1, C1 and C2, as well as between C2 and C3; the width of the mesophyle in C1, C2, and C3; the thickness of epidermal cells between the vascular bundle in C3 and the thickness of epidermal cells above the vascular bundles in C3. Pedicel length and width was measured with a Zeiss EVO 40 scanning electron microscope at an accelerating voltage of 15 kV.
One out of all investigated Typha glauca specimens produced seeds. Thirty seeds were collected from this specimen. Similarly 30 seeds were collected from one specimen of Typha angustifolia and one specimen of Typha latifolia each. Length and width of seeds were measured with the use of a stereoscopic microscope with a scale. Sculpture observations were conducted with a Zeiss EVO 40 scanning electron microscope at an accelerating voltage of 15 kV. As only one of 30 Typha glauca seeds showed a fully developed seed sculpture, SEM observations were limited to this very seed, as well as one other randomly selected seed of Typha angustifolia and Typha latifolia each. The measurements also included the length and width of 30 testa cells. Considering the fact that the compared samples were collected from single seeds, these observations should be treated as a preliminary study.
There have been no studies of seed sculpture in Typha sp. so far. Our preliminary observations showed, however that testa cells size may be a good diagnostic characteristics. We verified these conjectures with respect to parental species on a much larger sample measuring random testa cells in each 100 seeds of Typha angustifolia and Typha latifolia.
Calculations and statistical analyses of morpho-anatomical measurements
We used the hybrid index (Anderson, 1953) to verify the status of plants recognized in the field as pure species and hybrids. They began by testing the macro-morphological traits of Typha angustifolia and Typha latifolia with a Student’s t test to select traits, which discriminate both parents. Ten traits were found to strongly differentiate between Typha angustifolia and Typha latifolia (P < 0.001)—these were used to calculate the hybrid index: 1.—shoot diameter; 2 and 3—width of the 1st and 4th leaf; 4—leaf protrusion above the stem, 5 and 6—female spike length and width; 7—spike interval; 8—diameter of inflorescence rachis with pedicels; 9—inflorescence rachis diameter; and 10—plume length. The traits were divided into three classes: those ranging from the minimum value to 1/3 of the maximum value; those ranging from 1/3 to 2/3 of the maximum value; and those ranging from 2/3 of the maximum value to the maximum value (Pacheco et al., 1991). Class zero was defined as similar to Typha latifolia, class one as intermediate and class two as similar to Typha angustifolia.
We used a one-way ANOVA test followed by a Tukey’s post hoc test and a Chi square test (for bivariate data) to find, which of the 41 traits significantly differentiated the genetically identified cattail species. As we wanted to identify the best diagnostic traits for pure Typha glauca identification, we tested parents and hybrid F1 specimens.
Discriminant function analysis (DFA) was used to assess multivariate morphological differentiation between parents and Typha glauca and compare the potential for hybrid discrimination on the basis of morphological traits (macro-morphological data and pedicels data), as well as leaf anatomy. Two independent models were constructed for each of the groups of characteristics. Analyses were performed on the total set of 52 genetically identified individuals. Parental species were represented by 20 randomized specimens (listed in the “Measurements” section), whereas Typha glauca was represented by all F1 specimens. All analyzed variables from the study group were used in the model. To assess the impact of particular variables on the discriminant function, use was made of standardized coefficients (Barcikowski & Stevens, 1975). Seeds were not included in calculations due to the fact that measurements were not taken for particular individuals (see the “Measurements” section). ANOVA, as well as DFA were carried out by means of the Statistica software package (version 10, StatSoft).
In addition to macro-morphological traits, ANOVA with Tukey’s tests, as well as DFA analyses identified seven anatomical and micro-morphological characteristics as being indicative of the three examined taxa. We verified the diagnostic usefulness of these characteristics using the Anderson hybrid index. The index was tested extensively before the final scheme was accepted.
Additionally, we tested whether testa cells’ length and width are different for Typha latifolia and Typha angustifolia by means of Student’s t test.
Genetic analysis
Morphological data are likely to be unreliable in distinguishing F1 hybrids from their parents or backcrosses. Neither will such data indicate whether the parents are free of introgression. Therefore, we performed a genetic analysis of all 83 plants (30 individuals of the presumed “pure” T. latifolia, 28 individuals of “pure” T. angustifolia, and 26 individuals of their putative hybrids) using the randomly amplified polymorphic DNA (RAPD) by the methods of Williams et al. (1990) and Welsh and McClelland (1990).
The total genomic DNA was extracted from fresh and dried leaf tissue using a slightly modified cetyl trimethylammonium bromide (CTAB) method (Doyle & Doyle, 1987). DNA quality and concentration was estimated by electrophoresis and with a spectrophotometer, after which it was adjusted to 20 ng/μl and used as a template in PCR reactions. PCR RAPD amplifications were performed in 16 μl volumes containing a ready-to use PCR Master Mix (2x) (Fermentas), 20 pM 10-mer primer, and 50 ng of the template DNA. DNA amplification reactions were performed in a thermocycler (M. J. Research, Inc.) programmed for 40 cycles divided into two stages, differing in terms of annealing temperature as mentioned earlier (Czarna et al., 2013).
Initially, sixty 10-nucleotide primers (commercially available primer kits A, B and H, Operon Technologies) were probed for the preliminary selection of the most informative RAPD primers for the species in question. The bulked segregate analysis (BSA) technique (Michelmore et al., 1991) was used to identify primers producing clear RAPD markers that differentiate between Typha species. To determine differentiation among the analyzed plants and estimate the likelihood of occurrence of putative hybrid individuals among them, the initial reactions were conducted for all plants with 14 of the 21 primers chosen in this manner.
For the qualitative identification of hybrid individuals, diagnostic markers were defined. These markers are those present in all individuals of one species and absent in another, while species-specific markers are those that are unique to one species, but not necessarily present in all individuals within that species (Howard et al., 1997). To determine diagnostic markers, it is necessary to obtain "pure" samples of Typha latifolia and Typha angustifolia. Among the plants that contained morphologically extreme traits, type specimens were chosen for both Typha latifolia and Typha angustifolia and used to identify molecular markers specific to each species. To ensure that these markers reflected species-specific rather than site-specific differences, the specimens to be examined were sourced from different locations. Species-type specimens for Typha latifolia consisted of nine samples from different sites. The species standard for Typha angustifolia consisted of eight samples. All reference samples of parental species were screened using 21 RAPD primers, which previously seemed to differentiate between them. Nine of the selected primers produced clear reproducible RAPD fragments that contained diagnostic and species-specific markers. Finally, the DNA of 83 individuals were tested using only the nine primers that produce 20 diagnostic markers (eight for T. latifolia: OPA-01(2), OPA-11, OPA-13, OPA-15 (2), OPB-6 and OPB-12, and 12 for T. angustifolia: OPA-01, OPA-02, OPA-11, OPA-13, OPA-15, OPA-19 (2), OPB-6 (2), OPB-12 (2), and OPH-08).
Reaction products were separated by electrophoresis in 1.5% agarose gels in a 1xTBE buffer containing ethidium bromide in the presence of size markers. DNA bands were photographed in ultraviolet light utilizing the GBOX (Syngene Biotech) photo documentation system.
Calculations and statistical analyses for molecular data
Cluster analysis of the RAPD data obtained with 14 primers was conducted using the Gene Tools and Gene Directory software from Syngene Biotech.
The parentage of each individual sample was estimated using a hybrid index. Each plant was scored for the presence or absence of each parental (diagnostic) marker. Arithmetic hybrid index values (distances from Typha latifolia) were calculated as described in Fritz et al. (1994). Such calculations were as follows: “pure” Typha latifolia were given a score of 0; the presence of any Typha angustifolia marker or the lack of any Typha latifolia marker increased the index value up to a maximum of 1 for “pure” Typha angustifolia. F1 plants would theoretically have an index of 0.5 and possess all markers from both parents. Backcrosses are expected to lack a portion of the markers from one species. The presence of unique markers from both parents identifies the presence of a hybrid.
To investigate whether any correlation exists between the molecular hybrid index and the morphological hybrid index, use was made of the Spearmans’ rho method for abnormally distributed data.
Results
Morphologically intermediate cattails
Ten of the 14 macro-morphological traits were used to estimate the status of putative hybrids. The selected traits very significantly (P < 0.001) differentiated the specimens identified in the field as Typha angustifolia from those identified as Typha latifolia when subjected to the t test. The computed hybrid index values ranged from 0 to 20 and clearly separated the two parental species (Fig. 2a). Scores between 1 and 10 represent specimens of Typha latifolia, whereas scores between 14 and 19 represent specimens of Typha angustifolia. All individuals identified in the field as putative hybrids, except T-70, ended up aggregated in one group. While 10 specimens took intermediate hybrid index values (scores 11–13), 10 specimens fell well within the hybrid index distribution range of Typha latifolia (score 9) and Typha angustifolia (scores 14–17). Most of the intermediate individuals were collected in the Pomerania Region (samples T-71 to T-78) and are most likely to represent generation F1. The majority of the specimens having the hybrid index values that overlapped with those of Typha angustifolia originated from Mideast Poland (the Regions of Wielkopolska and Lower Silesia) and may be backcrosses or misidentified parents.
Frequency distribution of Typha latifolia, Typha angustifolia and Typha glauca individuals versus the hybrid index (HI) derived from 10 macro-morphological characteristics (a, b) and including the two new characteristics, i.e., length of pedicels and thickness of epidermal cells above vascular bundles (c). Histograms present individuals identified in the field (a) and genetically identified Typha spp (b, c). Stars indicate plants, which had been misidentified in the field. Striped areas show the distance between HI of Typha latifolia and probable F1 hybrids. Arrows indicate overlap of HI of probable F1 hybrids and Typha angustifolia
Genetic analysis (RAPDs) of Typha glauca and parental species
Initially, a total of 26 individuals of putative Typha glauca were examined and compared to both of the parental species using 14 primers that showed clear, reproducible products and seemed well differentiate between Typha angustifolia and Typha latifolia. All of these individuals showed such segregation of the species-specific markers that was observed in both parental species, not however, in equal proportions. In some of the cases, a greater similarity was observed with one of the parents, and in other cases with the other. On the other hand, clear differences in band patterns were observed in comparing the hybrid samples from Wielkopolska and Lower Silesia with those from other sampling sites. Moreover, the differentiation in band patterns observed among those hybrids was also greater. Additionally, analysis of profiles indicated that the differentiation level between individuals within each parental group was rather low. The genetic diversity of Typha latifolia was much lower than that of the Typha angustifolia and Typha glauca groups. A total of 116 scoreable RAPD markers, 111 of which were polymorphic, were evaluated in the range of between 200 and 1200 base pairs. An average of 8 polymorphic bands was amplified with each primer (the 5–12 range). Eighty-three out of 116 scored RAPD markers in Typha angustifolia were polymorphic (71.55%). In contrast, only 69 of the 116 scored markers in Typha latifolia were polymorphic (59.48%). Compared to them, the greatest number of polymorphic bands, with 101 of the 116 scored markers (87.07%), was observed in Typha glauca.
Cluster analysis of RAPD data allowed to group the plants in six main clusters. Two of them comprised Typha angustifolia samples, another two with Typha glauca ones, while yet another two included Typha latifolia. The latter is divided into four discrete groups, namely one comprising three Typha glauca individuals, one with many Typha latifolia and another two groups containing single putative hybrid individuals. Moreover, the Typha glauca accessions from Wielkopolska and Lower Silesia, as well as those from Pomerania and the herbarium, formed separate groups (Fig. 3). The dendrogram confirmed previous assumptions derived from genetic profile analysis whereby plants referred to as Typha glauca differ from the other examined species and, as such, are likely to constitute their hybrids.
Identification of RAPD species-specific markers
Results of the studies by other researchers, as well as our preliminary analysis indicated that RAPDs are powerful tools for characterizing hybrids between Typha species. Of the 60 primers tested in the study, we detected numerous markers that are unique for each parent. However, some of them were polymorphic within species, they are of limited use for quantifying the genetic composition of putative hybrids. Nine primers that produce 20 unambiguous marker loci were chosen for the purposes of this preliminary study. Eight of them are present in “pure” Typha latifolia and absent in “pure” Typha angustifolia while 12 are present in “pure” Typha angustifolia and absent in “pure” Typha latifolia. Species-diagnostic markers were used to identify hybrids between parental Typha species (Supplementary material 2). Based on these markers, 83 plants sourced from various locations across Poland were given hybrid index scores and plotted on Fig. 4. RAPD analysis confirmed the field identification of 11 plants as “pure” Typha latifolia—these were uniform with respect to the 20 genetic markers. Another nine plants (index = 0.05) differed only in that they lacked one of the eight Typha latifolia markers, or had a single Typha angustifolia marker, but were otherwise identifiable as Typha latifolia. Two further groups, which consisted of seven individuals in this cluster (0.1–0.15), represented nearly pure parental plants. Thirteen plants identified in the field as Typha angustifolia also appeared to be pure based on RAPDs (index = 1). Similarly as in the above case, 12 other plants differed only in that they lacked one of the 12 Typha angustifolia markers. Yet, neither of them had Typha latifolia markers (0.95). Together with plants T-37 and T-44, which were found in the field to be hybrids (0.85–0.9), they were classified as pure or nearly pure Typha angustifolia. Among the putative hybrids, four individuals exhibited perfect marker additivity as expected in an F1 (index = 0.5). Six other plants deviated by only one character (a distance of 0.45–0.55 from Typha latifolia), and were thus interpreted as F1-types. Another group (of six plants) deviated from F1 by only two characters (0.4–0.6) and were also classified as F1-types (Fig. 4). Among them was a plant (T-42) identified in the field as Typha angustifolia but appeared to be F1 based on RAPDs. Another plant (T-69), also misidentified in the field as Typha angustifolia, as well as an individual T-16 marked as Typha latifolia, appeared to belong to group of nine probably backcrosses toward Typha angustifolia (0.65–0.8). For the purposes of further analysis, these three plants are considered to be hybrids. Finally, a small cluster interpreted as a probable backcrosses toward Typha latifolia was created by four plants with a distance of 0.2–0.35 away from Typha latifolia.
Frequency distribution of individuals versus the hybrid index derived from the RAPD band data. The numbers of the individual plants are given; stars indicate plants which had been misidentified in the field. The figure shows clusters of the pure or nearly pure parents (index = 0 or 1), a cluster of probable F1 hybrids (index = 0.4–0.6), and probable backcrosses to Typha latifolia (index = 0.2–0.35) or to Typha angustifolia (index = 0.65–0.8)
Spearman’s rho showed that the distribution of cattail specimens according to the morphological hybrid index and molecular hybrid index were significantly correlated (ρ = −0.89, P < 0.00001).
Morphological and anatomical comparisons of genetically identified parents
Twenty-one of the 41 examined characteristics in genetically identified Typha angustifolia differed significantly from those observed in genetically identified Typha latifolia (Table 1). Insignificant results were obtained for three of 14 macro-morphological traits (shoot height, 1st leaf length, male spike length), pedicel width, two of the four characteristics of seeds (seed width, testa cell width), and 14 of the 21 the leaf anatomy characteristics (MC length; C1 width; no. of vascular bundles in C1; all measurements of C2; all measurements of C3; mesophyle width in C1, C2, and C3). Four characteristics (4th leaf width, shoot diameter, diameter of the inflorescence rachis with pedicels, and pedicels length) were clearly separated without an overlap between the parental species.
Morphological and anatomical analysis for Typha glauca (F1specimens) and parents
All 41 of the characteristics in question overlapped between Typha glauca and its parents. Hybrid specimens were intermediates between the parental plants for about half of the measured characteristics. The shares of intermediate characteristics were quite similar in the three trait sets (Table 1).
The variability coefficients (CVs) of 19 analyzed traits were higher in Typha glauca as compared to parental species. Such traits, as length of female spike, length of male spike, C2 length and C1 width were clearly more variable in hybrid as compared to parents. On the other hand, leaf protrusion above the stem was clearly less variable. Hybrid showed the variability coefficient CV = 0 for the number of vascular bundles in the margin compartment.
Twenty-one characteristics significantly differentiated hybrids from parental species (Tukey post hoc test, P < 0.05; see Table 1). Twelve traits differentiated hybrids from Typha angustifolia, while 16 traits differentiated them from Typha latifolia. The traits that differentiated Typha glauca and both parents included four macro-morphological characteristics (shoot length, 4th leaf width, female spike width, as well as the diameter of the inflorescence rachis with pedicels), pedicel length, length of cells in seed sculpture, and epidermal cell thickness located above vascular bundles. Additionally, the hybrid differed from Typha angustifolia in spike interval, seed length and width, MC width, and the number of vascular bundles in MC. The traits that exclusively differentiated the hybrid and Typha latifolia were six macro-morphological characteristics (stem diameter, 1st leaf width, female spike length, male spike length, diameter of the inflorescence rachis, and plume length) and three leaf anatomy traits (C1 length, width of beam between C1/C2, width of beam between C2/C3, and the epidermal cell thickness located between vascular bundles in C3).
Discriminant function analysis was prepared separately for morphological traits (macro-morphological traits and pedicels) and leaf anatomy. The hybrids were between parents in both of the obtained models (Fig. 5). In the model constructed for morphological data, the DF1 axis clearly discriminated Typha latifolia and was determined mostly by pedicel length and pedicel width (Table 2). The main species distinguished by the DF1 axis in models, which included leaf anatomy was Typha angustifolia. The epidermal cell thickness located above vascular bundles in C3 as well C3 length served as the most critical distinguishing factors. The second discrimination axis proved to have a relatively slight discriminatory power but was nevertheless statistically significant for morphology and anatomy. In both of the obtained models, DF2 primarily discriminated Typha glauca from parental plants. Pedicel width and spike interval (in the morphological model), as well as C2 length and C1 width (in the anatomy model) exerted the greatest influence on DF2.
ANOVA with Tukey’s tests, as well as discriminant function analyses showed that apart from the commonly used macro-morphological characteristics, there were some new traits, which could be helpful in identifying the cattails in question. Among them there are seven characteristics that are intermediate in hybrid (pedicel length and width, width of C1, width of beam between C1 and C2, width of beam between C2 and C3, and thickness of the epidermal cell located above/between vascular bundles). We tested all these characteristics using the Anderson hybrid index. Pedicel length and epidermal cell thickness located above vascular bundles proved to be the best discriminants. A comparison of hybrid indices constructed on the basis of macro-morphological traits with the hybrid index that included two new characteristics in addition to macro-morphological traits, was performed in Fig. 2b, c. In the first hybrid index, three F1 specimens overlapped with Typha angustifolia and none overlapped with Typha latifolia. After the addition of new characteristics, two F1 specimen overlapped with Typha angustifolia, whereas distance between Typha glauca and Typha latifolia was slightly increased.
Measurements of seed testa cells of Typha angustifolia and Typha latifolia by means of a scanning electron microscope (SEM) showed highly significant differences in epidermal cell length and width (Table 1). Typha glauca differed from parents as far as testa cell length is concerned. Epidermic cells were most frequently tetragonal with straight walls. In the case of Typha angustifolia, their length was two to three times greater than their width. In the case of Typha latifolia, it was roughly four to seven times greater, while in the case of Typha glauca, it turned out to be three to six times greater. The periclinal walls of epidermal cells were strongly depressed in each of the three species, while the cell borders were invisible. They were covered by a thin removable layer, which was easily torn. Anticlinal walls in all species were strongly raised (see Supplementary material 3).
Discussion
Reports of hybridization occurring between Typha latifolia and Typha angustifolia are common in the literature, especially in the USA and Canada. No reports, however, are available on hybrid plants between Typha latifolia and Typha angustifolia growing in central Europe. Two independent tests for evidence of the presence Typha glauca in Poland were used in this study: these were a nuclear genetic variation based on RAPD analysis and a morpho-anatomical variation. Also investigated was the possibility of backcrossing and genetic introgression in natural populations.
Occurrence of Typha glauca and parental species
The data collected during this study enabled us to identify three Typha glauca locations in north-western Poland (the Pomerania Region) and three Typha glauca locations in midwestern Poland (the Wielkopolska and Lower Silesia Regions). We also found herbarium specimens of the hybrid collected in five locations in southern Poland. The occurrence of hybrids in different and distant areas proves that hybridization in Poland is not a unique case. Each of the field-collected hybrid specimens grew alongside their parental species, similarly as in other regions of the world (Travis et al., 2011; Ball & Freeland, 2013). The hybrids were found in reservoirs characterized by highly variable water depths. These were mainly fish-breeding and country ponds, as well as backyard ponds and regulated river estuaries with concrete banks. These types of reservoirs, as well as (broadly speaking) disturbed sites, have been indicated as the sites of Typha glauca in both Eastern Europe (Kapitonova et al., 2012) and North America (Grace & Harrison, 1986; Olson et al., 2009).
Morphological and anatomical analysis for hybrids and parental plants
The morphological overlap between Typha glauca and parents has been well documented in North America (Smith, 1967). In studying Central European specimens, we demonstrated, in principle, the lack of discontinuity between species in all of the investigated macro-morphological traits. We also found that other traits (mostly those, which have not been previously investigated or studied as qualitative characteristics), such as leaf anatomy, pedicel size, as well as seed sizes and sculpture, overlapped with those of hybrid and parental plants. Where discontinuity between species was lacking, trait variability played a major role. Interspecific F1 hybrids were quite uniform and their general variation was very similar to that observed in the parents. As for the macro-morphological traits, the variability of Typha glauca tended to be only slightly higher than in the case of Typha angustifolia and Typha latifolia. These differences, however, did not seem to be as great as those observed by Kuehn et al. (1999) and Olson et al. (2009). In terms of anatomical traits, hybrids, as well as Typha angustifolia were noticeably more diverse than Typha latifolia.
We found that the certain sets of intermediate characteristics made hybrids resemble one of the parents more than the other. The F1 specimens in question were visibly closer to Typha angustifolia than to Typha latifolia. Similarities between hybrids and Typha angustifolia in terms of leaf and spike width were also shown by Kuehn et al. (1999) who used the bivariate scatterplot method. Polish hybrids bore a slight resemblance to the narrow-leaved cattail with their generative and vegetative body (e.g., small shoot diameter, narrow 1st leaf, small diameter of the inflorescence rachis, relatively short spikes, and short plume) and some anatomical features (mainly narrow beams between compartments). As far as spike interval length, seed size and some traits of margin compartments are concerned, the hybrids were relatively more similar to Typha latifolia.
The most discriminating characteristics were those positioned clearly in between parents and those which went beyond the values observed in parental species. Our preliminary identification, based solely on macro-morphological characteristics, was correct in about 90% of the cases. However, further tests showed that the diagnostic traits, which were substantially superior for hybrid identification were pedicel size and the size of epidermal cells in leaves. Pedicels of F1 hybrids (see Supplementary material 4) were slightly thicker than those of both parental plants (an average of 0.4 mm) and intermediate in their length (an average of 1 mm) ranging between the very short (an average of 0.5 mm) pedicels of Typha angustifolia and the long pedicels of Typha latifolia (an average of 1.5 mm). Epidermal cell thickness above vascular bundles in leaf anatomy (an average of 23 μm) always falls in between the narrower Typha angustifolia cells (an average of 18 μm) and the thicker Typha latifolia cells (an average of 30 μm), as shown in Supplementary material 5. The enlarged epidermis of Typha latifolia and Typha glauca was shown by McManus et al. (2002). They did not, however, point to the difference between the thickened cells of the hybrid and Typha latifolia. Marsh (1962), in his turn, failed to notice such differences at all. We also suggest that same other traits could be helpful in identifying species. Among them are compartment length (which is, two-and-a-half times longer than their width in Typha glauca, three times longer than their width in Typha latifolia, and roughly twice as long as their width in Typha angustifolia), as well as the number of vascular bundles in MC (all specimens of the hybrid sample showed one vascular bundle in the margin compartment, whereas not all parents had vascular bundles in their MCs).
In the course of this study, we compared seed size and sculpture for the three taxa. Size of hybrid seeds fell in between the sizes of their parents’ seeds, as in the Marsh study (1962). Testa cell size and shape were only observed in fully developed seeds—it revealed substantial differences between analyzed species. Our observations showed that length and width of testa cells strongly discriminated Typha angustifolia and Typha latifolia. They also suggest that these characteristics (in particular cell length) could be good diagnostic traits also for Typha glauca. However, in the case of Typha glauca, more hybrid seeds should be processed before a more resolute view on this issue can be expressed.
Genetic analysis of hybrid and parental DNA using RAPD
Cattail hybridization studies based on morphological, isozyme, and even molecular analyses have produced contradictory results (see introduction). On the other hand, the question of natural hybridization is vital not only for correct taxonomy but also due to its broader implications. Hybridization has been proposed as a likely driver of evolution. Hybrids may be more fit than their parent taxa thanks to such mechanisms as evolutionary novelty and greater genetic variation (Ellstrand & Schierenbeck, 2000). Compared to the above-mentioned studies, our study shows that band patterns characteristic of putative hybrids comprise the majority, but not all, of the markers found in parents. This study relied on molecular genetic markers (RAPDs) to, firstly, find whether the hybrid lineage (Typha glauca) is present in Poland, and secondly, to find whether any putative backcrossing occurs between them.
A preliminary analysis of the band profiles obtained for all Typha individuals showed low diversity observed especially for Typha latifolia (59.48%) as compared to Typha angustifolia (71.55%). This observation should be revised by analyzing more individuals and individuals from different sites, although it confirmed similar reports coming from the other regions described before. In studies of foreign geographic regions, Typha latifolia commonly showed a lower genetic variation than other Typha species, especially Typha angustifolia (Lee, 1975; Lamote et al., 2005; Tsyusko et al., 2005). The genetic diversity difference between Typha latifolia and Typha angustifolia was shown to be related to their different reproductive characteristics (Lee, 1975; Keane et al., 1999; Lamote et al., 2005; Tsyusko et al., 2005). Despite the low degree of diversity, a significant level of genetic differentiation was found between the Typha latifolia samples originating from different river basins. Whether this differentiation has any ecological relevance remains to be investigated (Lamote et al., 2005).
An analysis of a dendrogram obtained on the basis of polymorphic products generated by 14 RAPD primers produced data which are generally consistent with those obtained subsequently by calculating the hybrid index. Pure individuals were grouped into 4 separate clusters. Typha angustifolia created one big cluster comprised of pure specimens (an index close to 1) and one smaller cluster together with two misidentified plants (T-69, T-42) whose distance was nevertheless the longest in the group. A similar pattern was observed in Typha latifolia, where a smaller cluster was divided into three groups: one consisting of Typha latifolia and another two small comprising a few Typha glauca individuals classified as backcrosses in further analysis. A clear diversity was observed among hybrid plants from the Regions of Wielkopolska and Silesia which also differ from the Typha glauca found in other locations. An analysis of the dendrogram confirmed this observation. Both groups of Typha glauca created separate clusters, one of which was mostly comprised of hybrids subsequently classified as backcrosses. We assumed that the clustering based on polymorphic markers, which also included a set of diagnostic markers, grouped hybrids together separating them from pure species without differentiating the hybrids internally. This may well explain the presence of single backcrosses in the parental group clusters. Lamote et al. (2005) investigated Typha latifolia and Typha angustifolia collected in Flanders (North Belgium) by means of AFLP markers. In their studies, field determinations were also consistent with the subdivision of the clusters obtained on the basis of AFLP data. Typha latifolia samples formed a compact cluster while those of the Typha angustifolia were divided into smaller groups. They proposed different explanations which may lay the foundation of this subdivision: (1) the subdivision is a reflection of the higher diversity level expected in this species due to higher levels of cross-pollination, in comparison with Typha latifolia (Krattinger, 1975) and (2) one of the clusters corresponds to Typha glauca samples. Our observation seems to confirm that both of these explanations are reasonable.
Intermediate phenotypes are not always a reliable indicator of hybridization. To confirm that morphologically intermediate plants were indeed interspecific hybrids, RAPD markers were used to characterize the genotypic composition of a subset of morphologically intermediate plants. Parental species could be distinguished from one another based on 20 developed diagnostic markers. Generally speaking, the identification of species based on genetic data corresponded well with the identifications of species based on leaf width, which is a key morphological trait for distinguishing the two parental species. Almost all of the plants identified in the field as hybrids were confirmed to be indeed that through RAPD analysis. Only one plant (T-44) classified in the field as a hybrid was found to be Typha angustifolia. However, hybrids were found to be heterogeneous in their collection of parental markers. Although only four plants had the kind of complete additivity of markers one would expected to find in an F1, many of the hybrids had the majority of the 20 parental markers and were classified as F1-types. RAPD data also revealed evidence of hybridization that were not suspected based on morphology. Two plants considered to be Typha angustifolia were found in genetic analysis to be F1 hybrids (T-42) or a likely backcross to Typha angustifolia (T-69). On the other hand, three individuals: T-16, as well as T-2 together with T-40, were classified as Typha latifolia in the field, after which the RAPDs proved them to be backcrosses to Typha angustifolia and Typha latifolia respectively. This appears to illustrate a segregation of the few morphological traits that distinguish the species and indicates that morphology may not necessarily reflect the genetic contribution of the two parental species.
Backcrossed plants (which were generally in between hybrids and parent taxa) could be confused with F1 hybrids. F1 hybrids do not usually produce seeds or backcross to the parental species and are considered to be highly sterile. However, evidence of backcrossing in Ontario (Kirk et al., 2011) and Michigan (Snow et al., 2010) indicates that hybrids are fertile and that hybrid populations can include substantial numbers of backcrossed genotypes. As described by Snow et al. (2010), most of the backcrossed/advanced generation plants were more similar to Typha angustifolia, while a few were more related to Typha latifolia. This pattern suggests that crosses between F1 hybrids and each parent species are indeed possible.
In conclusion, RAPD analysis provided a measure of the nuclear composition of 29 hybrids and showed that 55% of all hybrids found may be classified as F1 with a hybrid index score 0.5 (4 individuals) or as F1-type with a hybrid value of 0.4–0.6 (12 individuals). Thirteen plants in the hybrid group had an index that was either lower or higher. This suggests that some of the hybrids are not F1 crosses but rather later generation backcrosses containing a high proportion of the Typha angustifolia or the Typha latifolia genome.
Our results confirmed the presence of hybrids in Poland and showed that, other than the F1-type, the analyzed Polish population of hybrids also includes 13 backcrossed genotypes, nine of them are similar to Typha angustifolia and four of them more similar to Typha latifolia.
To our knowledge, this paper is the first to describe the occurrence of hybrid generations and probably backcrossed generations in hybridizing cattail populations in this part of Europe.
References
Anderson, E., 1953. Introgressive hybridization. Biological Reviews 28: 280–307.
Angeloni, N. L., K. J. Jankowski, N. C. Tuchman & J. J. Kelly, 2006. Effects of an invasive cattail species (Typha × glauca) on sediment nitrogen and microbial community composition in a freshwater wetland. FEMS Microbiology Letters 263: 86–92.
Ball, D. & J. R. Freeland, 2013. Synchronous flowering times and asymmetrical hybridization in Typha latifolia and T. angustifolia in northeastern North America. Aquatic Botany 104: 224–227.
Barcikowski, R. S. & J. P. Stevens, 1975. A Monte Carlo study of the stability of canonical correlations, canonical weights and canonical variate–variable correlations. Multivariate Behavioral Research 10: 353–364.
Bayly, I. L. & T. A. O’Neill, 1971. A study of introgression in Typha at Point Pelee Marsh, Ontario. Canadian Field Naturalist 85: 309–314.
Carney, S. E., N. C. K. Gardner & L. H. Rieseberg, 2000. Evolutionary changes the fifty-year history of a hybrid population of sunflowers (Helianthus). Evolution 54: 462–474.
Clausen, K. E., 1962. Introgressive hybridization between two Minnesota Birches. Silvae Genetica 11: 142–150.
Corriveau, J. L. & A. W. Coleman, 1988. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany 75: 1443–1458.
Czarna, A., R. Nowińska & B. Gawrońska, 2013. Malus × oxysepala (M. domestica Borkh. × M. sylvestris Mill.)–a new spontaneous apple hybrid. Acta Societatis Botanicorum Poloniae 82(2): 147–156.
Doyle, J. J. & J. L. Doyle, 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Ellstrand, N. C. & K. A. Schierenbeck, 2000. Hybridization as a stimulus for the evolution of invasiveness in plants. Proceedings of the National Academy of Sciences of the United States of America 97: 7043–7050.
Farrell, J. M., B. A. Murry, D. J. Leopold, A. Halpern, M. B. Rippke, K. S. Godwin & S. D. Hafner, 2010. Water-level regulation and coastal wetland vegetation in the upper St. Lawrence River: inferences from historical aerial imagery, seed banks, and Typha dynamics. Hydrobiologia 647: 127–144.
Fassett, N. C. & L. L. Calhoun, 1952. Introgression between Typha latifolia and T. angustifolia. Evolution 6: 367–379.
Figert, E., 1890. Botanische Mitteilungen aus Schlesien. III. Typha latifolia × Typha angustifolia. Deutsche Botanische Monatsschrift 8: 55–57.
Fournier, P., 1961. Les quatre flores de la France. Lechevalier, Paris: 1106.
Fritz, R. S., C. M. Nichols-Orians & S. J. Brunsfeld, 1994. Interspecific hybridization of plants and resistance to herbivores: Hypotheses, genetics, and variable responses in a diverse herbivore community. Oecologia 97: 106–117.
Galatowitsch, S. M., N. O. Anderson & P. D. Ascher, 1999. Invasiveness in wetland plants in temperate North America. Wetlands 19: 733–755.
González-Rodrίguez, A., D. M. Arias, S. Valencia & K. Oyama, 2004. Morphological and RAPD analysis of hybridization between Quercus affinis and Q. laurina (Fagaceae), two Mexican red-oaks. American Journal of Botany 91: 401–409.
Govaerts, R., 2012. World Checklist of Typhaceae. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet; http://apps.kew.org/wcsp/ Retrieved 28.11.2012.
Grace, J. B. & J. S. Harrison, 1986. The biology of Canadian weeds. 73. Typha latifolia L., Typha angustifolia L. and Typha × glauca Godr. Canadian Journal of Plant Science 66: 361–379.
Howard, D. J., W. Preszler, J. Williams, S. Fenchel & W. J. Boecklen, 1997. How discrete are oaks species? Insights from a hybrid zone between Quercus grisea and Quercus gambelii. Evolution 51: 747–755.
Kapitonova, O. A., G. R. Platunova & V. I. Kapitonov, 2012. Rogozy Vjatsko-Kamskogo kraja. Izdatel’stvo Udmurtskij Universitet, Izhevsk: 191.
Keane, B., S. Pelikan, G. P. Toth, M. K. Smith & S. H. Rogstad, 1999. Genetic diversity of Typha latifolia (Typhaceae) and the impact of pollutants examined with tandem-repetitive DNA probes. American Journal of Botany 86: 1226–1238.
Kirk, H., C. Connelly & J. R. Freeland, 2011. Molecular genetic data reveal hybridization between Typha angustifolia and T. latifolia across a broad spatial scale in eastern North America. Aquatic Botany 95: 189–193.
Krattinger, K., 1975. Genetic mobility in Typha. Aquatic Botany 1: 57–70.
Kuehn, M. M. & B. N. White, 1999. Morphological analysis of genetically identified cattails Typha latifolia, Typha angustifolia, and Typha glauca. Canadian Journal of Botany 77: 906–912.
Kuehn, M. M., J. E. Minor & B. N. White, 1999. An examination of hybridization between the cattail species Typha latifolia and Typha angustifolia using random amplified polymorphic DNA and chloroplast DNA markers. Molecular Ecology 8: 1981–1990.
Lamont, B. B., T. He, N. J. Enright, S. L. Krauss & B. P. Miller, 2003. Anthropogenetic disturbance promotes hybridization between Banksia species by altering their biology. Journal of Evolutionary Biology 16: 551–557.
Lamote, V., M. De Loose, E. Van Bockstaele & I. Rolda′n-Ruiz, 2005. Evaluation of AFLP markers to reveal genetic diversity in Typha. Aquatic Botany 83: 296–309.
Larkin, D. J., M. J. Freyman, S. C. Lishawa, P. Geddes & N. C. Tuchman, 2012. Mechanisms of dominance by the invasive hybrid cattail Typha × glauca. Biological Invasions 14: 65–77.
Lee, D. W., 1975. Population variation and introgression in North American Typha. Taxon 24: 633–641.
Lishawa, S. C., D. A. Albert & N. C. Tuchman, 2010. Water level decline promotes Typha × glauca establishment and vegetation change in Great Lakes Coastal Wetlands. Wetlands 30: 1085–1096.
Luther, H., 1947. Typha angustifolia × latifolia L. (T. glauca Godr.) Ostfennoskandien. Memoranda Societatis Fauna Flora Fennica 23: 66–75.
Marsh, L. C., 1962. Studies in the genus Typha: I. Metaxenia, xenia and heterosis. II. Interspecific hybridization and the origin of T. glauca. III. Autecology, with special reference to the role of aerenchyma. PhD Thesis, Syracuse University, Syracuse, NY.
McManus, H. A., J. L. Seago Jr & L. C. Marsh, 2002. Epifluorescent and histochemical aspects of shoot anatomy of Typha latifolia L., Typha angustifolia L. and Typha glauca Godr. Annals of Botany 90: 489–493.
McNaughton, S. J., 1966. Ecotype function in the Typha community type. Ecological Monographs 36: 297–324.
Michelmore, R. W., I. Paran & R. V. Kessel, 1991. Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions using segregating populations. Proceedings of the National Academy of Sciences of the United States of America 88: 9828–9832.
Mirek, Z., H. Piękoś-Mirkowa, A. Zając & M. Zając, 2002. Flowering Plants and Pteridophytes of Poland. A Checklist. W. Szafer Institute of Botany, PAN, Kraków: 441 pp.
Olson, A., J. Paul & J. R. Freeland, 2009. Habitat preferencer of cattail species and hybrids (Typha spp.) in eastern Canada. Aquatic Botany 91: 67–70.
Pacheco, P., T. F. Stuessy & D. J. Crawford, 1991. Natural interspecific hybridization in Gunnera (Gunneraceae) of the Juan Fernandez Islands, Chile. Pacific Science 45: 389–399.
Rieseberg, L. H. & S. E. Carney, 1998. Plant hybridization. New Phytologist 140: 599–624.
Rieseberg, L. H., S. C. Kim, R. A. Randell, K. D. Whitney, B. L. Gross, C. Lexer & K. Clay, 2007. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129: 149–165.
Selbo, S. M. & A. A. Snow, 2004. The potential for hybridization between Typha angustifolia and Typha latifolia in a constructed wetland. Aquatic Botany 78: 361–369.
Sharitz, R. R., S. A. Wineriter, M. H. Smith & H. L. Edwin, 1980. Comparison of isozymes among Typha species in the eastern United States. American Journal of Botany 67: 1297–1303.
Shih, J. G. & S. A. Finkelstein, 2008. Range dynamics and invasive tendencies in Typha latifolia and Typha angustifolia in eastern North America derived from herbarium and pollen records. Wetlands 28: 1–16.
Smith, S. G., 1967. Experimental and natural hybrids in North American Typha (Typhaceae). The American Midland Naturalist 78: 257–287.
Smith, S. G., 1987. Typha: its taxonomy and the ecological significance of hybrids. Archiv fur Hydrobiologie 27: 129–138.
Smith, S. G., 2000. Typhaceae Flora of North America, Vol. 22. Oxford University Press, New York.
Snow, A. A., S. E. Travis, R. Wildova, T. Fer, P. M. Sweeney, J. E. Marburger, S. Windels, B. Kubatova, D. E. Goldberg & E. Mutegi, 2010. Species-specific SSR alleles for studies of hybrid cattails (Typha latifolia × T. angustifolia; Typhaceae) in North America. American Journal of Botany 97: 2061–2067.
Stace, C. A., 2005. New Flora of the British Isles. Cambridge University Press, Cambridge.
Tovar-Sánchez, E. & K. Oyama, 2004. Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: Morphological and molecular evidence. American Journal of Botany 91: 1352–1363.
Travis, S. E., J. E. Marburger, S. Windels & B. Kubátová, 2010. Hybridization dynamics of invasive cattail (Typhaceae) stands in the Western Great Lakes Region of North America: A molecular analysis. Journal of Ecology 98: 7–16.
Travis, S. E., J. E. Marburger, S. K. Windels & B. Kubátová, 2011. Clonal structure of invasive cattail (Typhaceae) stands in the Upper Midwest Region of the US. Wetlands 31: 221–228.
Tsyusko, O. V., M. H. Smith, R. R. Sharitz & T. C. Glenn, 2005. Genetic and clonal diversity of two cattail species, Typha latifolia and T. angustifolia (Typhaceae), from Ukraine. American Journal of Botany 92: 1161–1169.
Tuchman, N. C., D. J. Larkin, P. Geddes, R. Wildova, K. Jankowski & D. E. Goldberg, 2009. Patterns of environmental change associated with Typha × glauca invasion in a Great Lakes coastal wetlands. Wetlands 29: 964–975.
Waters, I. & J. M. Shay, 1990. A field study of the morphometric response of Typha glauca shoots to a water depth gradient. Canadian Journal of Botany 68: 2339–2343.
Waters, I. & J. M. Shay, 1991. Effect of water depth on population parameters of a Typha glauca stand. Canadian Journal of Botany 70: 349–351.
Welsh, J. & M. McClelland, 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 18: 7213–7218.
Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski & S. V. Tingey, 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18: 6531–6535.
Zedler, J. B. & S. Kercher, 2004. Causes and consequences of invasive plants in wetlands: Opportunities, opportunists, and outcomes. Critical Reviews in Plant Sciences 70: 431–452.
Acknowledgments
This research was supported by Ministry of Research and High Education Research Capacity Grants for Poznań Universiy of Life Sciences (Nos. 508.641.00 and 508.181.00). We would like to thank Ilona Wysakowska for her assistance with the laboratory work and Wojciech Wieczorek for his help in SEM measurements. We also gratefully acknowledge valuable comments of the anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: M. T. Ferreira, M. O’Hare, K. Szoszkiewicz & S. Hellsten / Plants in Hydrosystems: From Functional Ecology to Weed Research
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Nowińska, R., Gawrońska, B., Czarna, A. et al. Typha glauca Godron and its parental plants in Poland: taxonomic characteristics. Hydrobiologia 737, 163–181 (2014). https://doi.org/10.1007/s10750-014-1862-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-014-1862-0