Abstract
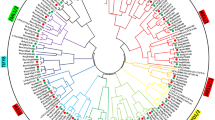
TIFY protein is a plant-specific transcription factor widely found in land plants and plays an important role in plant growth and development, signal transduction and stress response. Although many TIFY gene families have been identified and studied in plants, they have not been well described in octaploid strawberry. In this study, 54 TIFY family genes in strawberry were identified and their biological information was analyzed. The phylogenetic tree showed that FaTIFY genes were divided into four subfamilies: TIFY, JAZ, ZML and PPD. The JAZ subfamily was the largest and can be further divided into five groups. These genes are located on six chromosomes and exhibit a motif range of 3 to 8. Promoter cis-element analysis showed that most FaTIFY genes contain cis-elements associated with plant hormones responses. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed that FaTIFY genes had different expression patterns under methyl jasmonate (MeJA), and salicylic acid (SA) treatments. At the same time, we also examined the expression pattern of TIFY gene after boea mold infection and predicted the binding of TIFY protein to WRKY transcription factor, providing a theoretical basis for exploring the role of TIFY gene in hormone delivery and plant defense in strawberry.







Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
References
Aparicio-Fabre R, Guillén G, Loredo M et al (2013) Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol 13:26. https://doi.org/10.1186/1471-2229-13-26
Babalar M, Asghari M, Talaei A, Khosroshahi A (2007) Effect of pre- and postharvest salicylic acid treatment on ethylene production, fungal decay and overall quality of Selva Strawberry fruit. Food Chem 105:449–453. https://doi.org/10.1016/j.foodchem.2007.03.021
Bailey TL, Boden M, Buske FA et al (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Chen C, Chen H, Zhang Y et al (2020a) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chen L, Zhang L, Xiang S et al (2020b) The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J Exp Bot 72:1473–1489. https://doi.org/10.1093/jxb/eraa529
Chini A, Fonseca S, Fernández G et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666–671. https://doi.org/10.1038/nature06006
Cui X, Yan Q, Gan S et al (2019) GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol 19:598. https://doi.org/10.1186/s12870-019-2132-0
de Vries S, de Vries J, von Dahlen JK et al (2018) On plant defense signaling networks and early land plant evolution. Commun Integr Biol 11:1–14. https://doi.org/10.1080/19420889.2018.1486168
Demianski AJ, Chung KM, Kunkel BN (2012) Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol Plant Pathol 13:46–57. https://doi.org/10.1111/j.1364-3703.2011.00727.x
Edger PP, Poorten TJ, VanBuren R et al (2019) Origin and evolution of the octoploid strawberry genome. Nat Genet 51:541–547. https://doi.org/10.1038/s41588-019-0356-4
Galan AGA, Doll J, Saile SC et al (2023) The non-JAZ TIFY protein TIFY8 of Arabidopsis thaliana interacts with the HD-ZIP III transcription factor REVOLUTA and regulates leaf senescence. Int J Mol Sci 24:3079. https://doi.org/10.3390/ijms24043079
Garrido-Bigotes A, Figueroa NE, Figueroa PM, Figueroa CR (2018) Jasmonate signalling pathway in strawberry: genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening. PLoS ONE 13:e0197118. https://doi.org/10.1371/journal.pone.0197118
Gimenez-Ibanez S, Boter M, Ortigosa A et al (2016) JAZ 2 controls stomata dynamics during bacterial invasion. N Phytol 213:1378–1392. https://doi.org/10.1111/nph.14354
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Guo Q, Yoshida Y, Major IT et al (2018) JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc Natl Acad Sci 115:E10768–E10777. https://doi.org/10.1073/pnas.1811828115
Hancock JF, Finn CE, Luby JJ et al (2010) Reconstruction of the strawberry, Fragaria ×ananassa, using genotypes of F. Virginiana and F. Chiloensis. HortScience 45:1006–1013. https://doi.org/10.21273/hortsci.45.7.1006
Hardigan MA, Lorant A, Pincot DDA et al (2021) Unraveling the complex hybrid ancestry and domestication history of cultivated strawberry. Mol Biol Evol 38:2285–2305. https://doi.org/10.1093/molbev/msab024
Hu B, Jin J, Guo AY et al (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Jung S, Lee T, Cheng CH et al (2018) 15 years of GDR: New data and functionality in the genome database for Rosaceae. Nucleic Acids Res 47:D1137–D1145. https://doi.org/10.1093/nar/gky1000
Katsir L, Chung HS, Koo AJ, Howe GA (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11:428–435. https://doi.org/10.1016/j.pbi.2008.05.004
Lescot M, Déhais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Li X, Yin X, Wang H et al (2014) Genome-wide identification and analysis of the apple (Malus × Domestica Borkh.) TIFY gene family. Tree Genet Genomes 11:808. https://doi.org/10.1007/s11295-014-0808-z
Liu X, Yu F, Yang G et al (2022a) Identification of TIFY gene family in walnut and analysis of its expression under abiotic stresses. BMC Genom 23:190. https://doi.org/10.1186/s12864-022-08416-9
Liu YL, Zheng L, Jin LG et al (2022b) Genome-wide analysis of the soybean TIFY family and identification of GmTIFY10e and GmTIFY10g response to salt stress. Front Plant Sci 13:845314. https://doi.org/10.3389/fpls.2022.845314
Ma Y, Cao J, He J et al (2018) Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int J Mol Sci 19:3643. https://doi.org/10.3390/ijms19113643
Ma Y, Ran J, Li G et al (2023) Revealing the roles of the JAZ family in defense signaling and the agarwood formation process in Aquilaria sinensis. Int J Mol Sci 24:9872. https://doi.org/10.3390/ijms24129872
Moreno JE, Shyu C, Campos ML et al (2013) Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol 162:1006–1017. https://doi.org/10.1104/pp.113.218164
Muhae-Ud-Din G, Chen D, Liu T et al (2020) Methyljasmonate and salicylic acid contribute to the control of Tilletia Controversa Kühn, causal agent of wheat dwarf bunt. Sci Rep 10:19175. https://doi.org/10.1038/s41598-020-76210-2
Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23:3089–3100. https://doi.org/10.1105/tpc.111.089300
Pérez AC, Durand AN, Bossche RV et al (2014) The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS ONE 9:e84891. https://doi.org/10.1371/journal.pone.0084891
Ruan J, Zhou Y, Zhou M et al (2019) Jasmonic acid signaling pathway in plants. Int J Mol Sci 20:2479. https://doi.org/10.3390/ijms20102479
Saha G, Park JI, Kayum MA, Nou IS (2016) A genome-wide analysis reveals stress and hormone responsive patterns of TIFY family genes in Brassica rapa. Front Plant Sci 7:936. https://doi.org/10.3389/fpls.2016.00936
Shen J, Zou Z, Xing H et al (2020) Genome-wide analysis reveals stress and hormone responsive patterns of JAZ family genes in Camellia Sinensis. Int J Mol Sci 21:2433. https://doi.org/10.3390/ijms21072433
Shikata M, Matsuda Y, Ando K et al (2004) Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J Exp Bot 55:631–639. https://doi.org/10.1093/jxb/erh078
Singh P, Mukhopadhyay K (2021) Comprehensive molecular dissection of TIFY transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat (Triticum aestivum L). Sci Rep 11:9739. https://doi.org/10.1038/s41598-021-87722-w
Sun Y, Liu C, Liu Z et al (2021) Genome-wide identification, characterization and expression analysis of the JAZ gene family in resistance to gray leaf spots in tomato. Int J Mol Sci 22:9974. https://doi.org/10.3390/ijms22189974
Thatcher LF, Cevik V, Grant M et al (2016) Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J Exp Bot 67:2367–2386. https://doi.org/10.1093/jxb/erw040
Thines B, Katsir L, Melotto M et al (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448:661–665. https://doi.org/10.1038/nature05960
Thireault C, Shyu C, Yoshida Y et al (2015) Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J 82:669–679. https://doi.org/10.1111/tpj.12841
Tripathi D, Raikhy G, Kumar D (2019) Chemical elicitors of systemic acquired resistance—salicylic acid and its functional analogs. Curr Plant Biol 17:48–59. https://doi.org/10.1016/j.cpb.2019.03.002
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498. https://doi.org/10.1016/j.pbi.2004.07.012
Vanholme B, Grunewald W, Bateman A et al (2007) The tify family previously known as ZIM. Trends Plant Sci 12:239–244. https://doi.org/10.1016/j.tplants.2007.04.004
Wager A, Browse J (2012) Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Sci 3:41. https://doi.org/10.3389/fpls.2012.00041
Wang N, Xiang Y, Fang L et al (2013) Patterns of gene duplication and their contribution to expansion of gene families in grapevine. Plant Mol Biol Rep 31:852–861. https://doi.org/10.1007/s11105-013-0556-5
Wang Y, Pan F, Chen D et al (2017) Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol Biochem 115:360–371. https://doi.org/10.1016/j.plaphy.2017.04.015
Wang Y, Li Y, He SP et al (2019) A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol Biochem 141:231–239. https://doi.org/10.1016/j.plaphy.2019.06.005
Wang H, Leng X, Xu X, Li C (2020) Comprehensive analysis of the TIFY gene family and its expression profiles under phytohormone treatment and abiotic stresses in roots of Populus trichocarpa. Forests 11:315. https://doi.org/10.3390/f11030315
White DWR (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci 103:13238–13243. https://doi.org/10.1073/pnas.0604349103
Xia W, Yu H, Cao P et al (2017) Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front Plant Sci 8:493. https://doi.org/10.3389/fpls.2017.00493
Ye H, Du H, Tang N et al (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305. https://doi.org/10.1007/s11103-009-9524-8
Yocca AE, Edger PP (2022) Current status and future perspectives on the evolution of cis-regulatory elements in plants. Curr Opin Plant Biol 65:102139. https://doi.org/10.1016/j.pbi.2021.102139
Zabala M, de Zhai T, Jayaraman B S, et al (2016) Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. N Phytol 209:1120–1134. https://doi.org/10.1111/nph.13683
Zhang Y, Li X (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol 50:29–36. https://doi.org/10.1016/j.pbi.2019.02.004
Zhang Y, Gao M, Singer SD et al (2012) Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 7:e44465. https://doi.org/10.1371/journal.pone.0044465
Zhang C, Atanasov KE, Murillo E et al (2023) Spermine deficiency shifts the balance between jasmonic acid and salicylic acid-mediated defence responses in Arabidopsis. Plant Cell Environ 46:3949–3970. https://doi.org/10.1111/pce.14706
Zheng L, Wan Q, Wang H et al (2022) Genome-wide identification and expression of TIFY family in cassava (Manihot esculenta Crantz). Front Plant Sci 13:1017840. https://doi.org/10.3389/fpls.2022.1017840
Funding
This research was supported by the National Natural Science Foundation of China (32272370).
Author information
Authors and Affiliations
Contributions
Xingfeng Shao (XS), Yi Chen (YC), and Siyao Tong (ST) conceptualized and designed the experiments. ST conducted the experiments and drafted the manuscript. Data analysis was performed by ST, YC, and Yingying Wei (YW). The manuscript’s initial draft was critically reviewed and edited by XS, YC, YW, Jianfen Ye (JY), Shu Jiang (SJ), and Feng Xu (FX). Funding acquisition was managed by XS.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tong, S., Chen, Y., Wei, Y. et al. Genome-wide identification and response to exogenous hormones and pathogens of the TIFY gene family in Fragaria ananassa. Plant Growth Regul (2024). https://doi.org/10.1007/s10725-024-01147-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10725-024-01147-9