Abstract
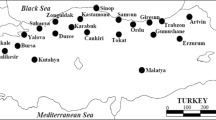
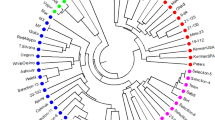
Camellia oleifera Abel., as one of the four major woody oilseeds, has a high economic value, and the wild C. oleifera genes, whose distribution area is located at the northern edge, are abundant and are valuable resources for C. oleifera breeding. In this study, a total of 341 wild C. oleifera populations were sampled from 11 different localitions in Xinxian County, the hinterland of the Dabie Mountains in the northern margin of the distribution of C. oleifera in China, and 16 pairs of simple sequence repeat (SSR) molecular markers were used to analyse the genetic diversity. Using these 16 pairs of primers to detect the genetic diversity of the wild C. oleifera population, 174 alleles were amplified. The average number of alleles (Na) was 10.875, the average expected heterozygosity (He) was 0.739, the observed heterozygosity (Ho) was 0.718, and the average polymorphic information index (PIC) was 0.739. The 11 wild C. oleifera populations in Xinxian County had high genetic diversity, and the average expected heterozygosity (He) among populations was 0.735. The molecular variance showed that the genetic variation mainly came from within the population, accounting for 88.21% of the total variation. The genetic differentiation coefficient Fst between populations was small, with an average of only 0.04. According to the results of Structure and principal cordinate analysis (PCoA) and cluster analysis, these 11 populations could be roughly divided into two categories. The Mantel test preferentially clustered some populations close to each other, but there was no significant correlation between genetic distance and geographical distance. We provides a theoretical basis for the rational development and utilisation of wild C. oleifera resources in the future and provide a scientific and technological method for future breeding.









Similar content being viewed by others
References
Bandupriya HDD, Perera SACN, Ranasinghe CS et al (2022) Physiological, biochemical and molecular evaluation of micropropagated and seed-grown coconut (Cocos nucifera L.) palms. Trees 36:127–138
Blacket MJ, Robin C, Good RT et al (2012) Universal primers for fluorescent labelling of PCR fragments—an efficient and cost-effective approach to genotyping by fluorescence. Mol Ecol Resour 12:456–463
Bruvo R, Michiels NK, D’Souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level: microsatellite distances in polyploids. Mol Ecol 13:2101–2106
Cavers S, Degen B, Caron H et al (2005) Optimal sampling strategy for estimation of spatial genetic structure in tree populations. Heredity 95:281–289
Chen J, Yang X, Huang X et al (2017) Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genom 18:211
Chen T, Liu L, Zhou Y et al (2023) Characterization and comprehensive evaluation of phenotypic characters in wild Camellia oleifera germplasm for conservation and breeding. Front Plant Sci 14:1052890
Chen Y, Dai X, Hou J et al (2016) DNA fingerprinting of oil camellia cultivars with SSR markers. Tree Genet Genom 12:7
Cui X, Li C, Qin S et al (2022a) High-throughput sequencing-based microsatellite genotyping for polyploids to resolve allele dosage uncertainty and improve analyses of genetic diversity, structure and differentiation: a case study of the hexaploid Camellia oleifera. Mol Ecol Resour 22:199–211
De Kort H, Prunier JG, Ducatez S et al (2021) Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nat Commun 12:516
Deng Q, Li J, Gao C et al (2020) New perspective for evaluating the main Camellia oleifera cultivars in China. Sci Rep 10:20676
Dong B, Deng Z, Liu W et al (2022) Development of expressed sequence tag simple sequence repeat (EST-SSR) markers and genetic resource analysis of tea oil plants (Camellia spp.). Conserv Genet Resour 14:41–45
Guney M, Kafkas S, Keles H et al (2021) Genetic diversity among some Walnut (Juglans regia L.) genotypes by SSR Markers. Sustainability 13:6830
He Z, Liu C, Wang X et al (2020) Assessment of genetic diversity in Camellia oleifera Abel. accessions using morphological traits and simple sequence repeat (SSR) markers. Breed Sci 70:586–593
Healey A, Furtado A, Cooper T, Henry RJ (2014) Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:21
Huang B, Wang Z, Huang J et al (2022) Population genetic structure analysis reveals significant genetic differentiation of the endemic species Camellia chekiangoleosa Hu. with a Narrow Geographic Range. Forests 13:234
Huang X (2016) Genetic structure ofhexaploid wild Camellia oleifera in mountJinggang and Lu based on microsatellite markers. Nanchang University, Master
Jia B, Lin Q, Zhang L et al (2014) Development of 15 genic-ssr markers in oil-tea tree (Camellia oleifera) based on transcriptome sequencing. Genetika 46:789–797
Karagöz H, Hosseinpour A, Karagöz FP et al (2022) Dissection of genetic diversity and population structure in oregano (Origanum acutidens L.) genotypes based on agro-morphological properties and start codon targeted (SCoT) markers. Biologia 77:1231–1247
Li H, Ding H, Chen Y et al (2017) ldentification of 12 superior cultivars of camellia oleifera by using simple sequence repeat feature lndexes. J Chin Cereals Oils 32:171–178
Lin P, Wang K, Wang Y et al (2022) The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol 23:14
Meirmans PG, Liu S (2018) Analysis of molecular variance (AMOVA) for Autopolyploids. Front Ecol Evol 6:66
Minh Nguyen D, Lan Phan Nguyen H, Minh Nguyen T (2022) Genetic structure of the endemic Dipterocarpus condorensis revealed by microsatellite markers. AoB Plants 14:plac007
Molloy EK, Durvasula A, Sankararaman S (2021) Advancing admixture graph estimation via maximum likelihood network orientation. Bioinformatics 37:i142–i150
Mousavi S, Stanzione V, Mencuccini M et al (2019) Biochemical and molecular profiling of unknown olive genotypes from central Italy: determination of major and minor components. Eur Food Res Technol 245:83–94
Özkan G, Haliloğlu K, Türkoğlu A et al (2022) Determining genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from Türkiye using SSR markers. Genes 13:1410
Öztürk Hİ, Dönderalp V, Bulut H, Korkut R (2022) Morphological and molecular characterization of some pumpkin (Cucurbita pepo L.) genotypes collected from Erzincan province of Turkey. Sci Rep 12:6814
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pometti CL, Bessega CF, Vilardi JC et al (2015) Genetic diversity within and among two Argentinean and one Mexican species of Acacia (Fabaceae). Bot J Linnean Soc 177:593–606
Qi H, Sun X, Yan W et al (2020) Genetic relationships and low diversity among the tea-oil Camellia species in Sect. Oleifera, a bulk woody oil crop in China. Front Plant Sci 13:996731
Stankiewicz KH, Vasquez Kuntz KL, Coral Microsatellite Group, Baums IB (2022) The impact of estimator choice: disagreement in clustering solutions across K estimators for Bayesian analysis of population genetic structure across a wide range of empirical data sets. Mol Ecoly Resour 22:1135–1148
Sütyemez M, Bükücü ŞB, Keleş Ö et al (2021) Phenological differences, genetic diversity, and population structure of genotypes obtained from seeds of Kaman-1 Walnut cultivar. J Food Qual 2021:1–9
Tarang A, Kordrostami M, Shahdi Kumleh A et al (2020) Study of genetic diversity in rice (Oryza sativa L.) cultivars of Central and Western Asia using microsatellite markers tightly linked to important quality and yield related traits. Genet Resour Crop Evol 67:1537–1550
Verhoeven KJF, vonHoldt BM, Sork VL (2016) Epigenetics in ecology and evolution: what we know and what we need to know. Mol Ecol 25:1631–1638
Vuletin Selak G, Baruca Arbeiter A, Cuevas J et al (2021) Seed paternity analysis using SSR markers to assess successful pollen donors in mixed olive orchards. Plants 10:2356
Wang F, Song F, Song M et al (2022) Genetic structure and paternal admixture of the modern Chinese Zhuang population based on 37 Y-STRs and 233 Y-SNPs. Foren Sci Int: Genet 58:102681
Yilmaz A, Ciftci V (2021) Genetic relationships and diversity analysis in Turkish laurel (Laurus nobilis L.) germplasm using ISSR and SCoT markers. Mol Biol Rep 48:4537–4547
Yin X, Li T, Tian QQ et al (2022) Development of novel polymorphic microsatellite markers and their application for closely related Camellia (Theaceae) species. Russ J Genet 58:404–412
Zhang M, Yao Y, Chen S (2016) Genetic diversity analysis of tea germplasm in Qiannan prefecture by SSR markers. Acta Botan Boreali-Occiden Sin 36:1117–1124
Zhang Z, Meng J, Pan D et al (2019) Mating system and progeny genetic diversity of Camellia oleifera ‘Ruan Zhi.’ J for Res 30:1805–1810
Zhao Y, Ruan CJ, Ding GJ, Mopper S (2017) Genetic relationships in a germplasm collection of Camellia japonica and Camellia oleifera using SSR analysis. Genet Mol Res 16. https://doi.org/10.4238/gmr16019526
Zhou B, Wang L, Xu X et al (2020) Analysis of genetic diversity and genetic relationship of Leibo wild tea resources. J Yunnan Agric Univ (nat Sci) 35:122–129
Zhou L, Wang X, Wang L et al (2015) Genetic diversity of oil-tea camellia germplasms revealed by ISSR analysis. Int J Biomath 08:1550070
Zhu Y, Liang D, Song Z et al (2022) Genetic diversity analysis and core germplasm collection construction of Camellia oleifera based on fruit phenotype and SSR data. Genes 13:2351
Funding
This research was funded by the project of National Natural Science Foundation of China, grant number “32060362” and Supported by the Jiangxi Forestry Bureau Camellia Research Project grant number “YCYJZX [2023]121”.
Author information
Authors and Affiliations
Contributions
Methodology, D.H. and J.L.; software, S.Y.; formal analysis, L.Y. and Y.Z.; investigation, L.C.andB.C.; data curation,L.C. and Q.C.; writing—original draft preparation, L.C.; writing—review and editing, L.C.; project administration, D.H.; funding acquisition, D.H. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any research with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongnan Hu have contributed equally to this work.
Appendix
Appendix
DNA concentration of single plant leaves within each wild Camellia oleifera population.
Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml |
|---|---|---|---|---|---|---|---|---|
KB | 1 | 116.7 | WW | 1 | 258.2 | TG | 1 | 70.4 |
2 | 119.0 | 2 | 96.9 | 2 | 63.5 | |||
3 | 109.3 | 3 | 54.1 | 3 | 80.7 | |||
4 | 117.0 | 4 | 73.5 | 4 | 99.0 | |||
5 | 105.5 | 5 | 126.8 | 5 | 72.7 | |||
6 | 149.2 | 6 | 68.0 | 6 | 70.0 | |||
7 | 167.4 | 7 | 80.0 | 7 | 72.4 | |||
8 | 58.8 | 8 | 77.6 | 8 | 116.2 | |||
9 | 115.2 | 9 | 55.6 | 9 | 89.3 | |||
10 | 61.5 | 10 | 94.2 | 10 | 66.7 | |||
11 | 80.4 | 11 | 245.3 | 11 | 82.9 | |||
12 | 37.4 | 12 | 74.0 | 12 | 83.7 | |||
13 | 70.5 | 13 | 97.1 | 13 | 61.8 | |||
14 | 90.3 | 14 | 65.6 | 14 | 67.8 | |||
15 | 88.9 | 15 | 200.2 | 15 | 62.7 | |||
16 | 82.2 | 16 | 139.2 | 16 | 79.3 | |||
17 | 83.5 | 17 | 95.7 | 17 | 46.6 | |||
18 | 66.6 | 18 | 74.1 | 18 | 42.2 | |||
19 | 70.5 | 19 | 132.1 | 19 | 63.7 | |||
20 | 57.5 | 20 | 103.6 | 20 | 72.0 | |||
21 | 55.6 | 21 | 96.6 | 21 | 93.2 | |||
22 | 75.7 | 22 | 81.6 | 22 | 47.4 | |||
23 | 129.2 | 23 | 86.0 | 23 | 106.6 | |||
24 | 105.9 | 24 | 54.8 | 24 | 54.5 | |||
25 | 77.6 | 25 | 87.2 | 25 | 63.2 | |||
26 | 84.7 | 26 | 124.9 | 26 | 51.1 | |||
27 | 59.2 | 27 | 74.1 | 27 | 57.7 | |||
28 | 55.4 | 28 | 56.6 | 28 | 57.4 | |||
29 | 71.5 | 29 | 43.7 | 29 | 70.4 | |||
30 | 95.4 | 30 | 87.1 | 30 | 67.4 | |||
31 | 33.2 | 31 | 96.0 | 31 | 76.7 | |||
Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml |
JLS | 1 | 30.7 | BLF | 1 | 68.4 | TF | 1 | 48.0 |
2 | 34.6 | 2 | 55.4 | 2 | 57.8 | |||
3 | 48.2 | 3 | 72.0 | 3 | 75.1 | |||
4 | 30.0 | 4 | 51.4 | 4 | 97.1 | |||
5 | 30.9 | 5 | 130.9 | 5 | 91.3 | |||
6 | 101.5 | 6 | 102.9 | 6 | 39.1 | |||
7 | 46.9 | 7 | 81.2 | 7 | 79.8 | |||
8 | 57.9 | 8 | 49.3 | 8 | 94.3 | |||
9 | 55.2 | 9 | 51.7 | 9 | 90.5 | |||
10 | 53.0 | 10 | 59.8 | 10 | 48.7 | |||
11 | 56.7 | 11 | 57.7 | 11 | 91.8 | |||
12 | 72.2 | 12 | 46.5 | 12 | 75.9 | |||
13 | 58.8 | 13 | 21.4 | 13 | 48.9 | |||
14 | 79.4 | 14 | 37.8 | 14 | 49.6 | |||
15 | 76.8 | 15 | 46.8 | 15 | 59.0 | |||
16 | 76.2 | 16 | 69.2 | 16 | 54.9 | |||
17 | 125.4 | 17 | 82.6 | 17 | 45.0 | |||
18 | 75.9 | 18 | 77.5 | 18 | 61.8 | |||
19 | 82.5 | 19 | 65.1 | 19 | 68.7 | |||
20 | 64.1 | 20 | 64.2 | 20 | 54.8 | |||
21 | 83.7 | 21 | 42.9 | 21 | 51.1 | |||
22 | 108.0 | 22 | 99.5 | 22 | 65.2 | |||
23 | 114.5 | 23 | 90.6 | 23 | 61.8 | |||
24 | 83.0 | 24 | 129.1 | 24 | 66.3 | |||
25 | 69.8 | 25 | 77.8 | 25 | 43.1 | |||
26 | 46.2 | 26 | 602 | 26 | 105.4 | |||
27 | 99.2 | 27 | 62.3 | 27 | 106.2 | |||
28 | 49.6 | 28 | 75.7 | 28 | 67.4 | |||
29 | 92.7 | 29 | 47.2 | 29 | 79.0 | |||
30 | 91.0 | 30 | 56.9 | 30 | 54.5 | |||
31 | 70.7 | 31 | 48.2 | 31 | 79.4 | |||
Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml |
JL | 1 | 45.6 | LQ | 1 | 37.7 | XH | 1 | 90.5 |
2 | 37.0 | 2 | 74.7 | 2 | 106.1 | |||
3 | 22.5 | 3 | 27.0 | 3 | 74.9 | |||
4 | 33.9 | 4 | 68.3 | 4 | 34.0 | |||
5 | 65.9 | 5 | 54.1 | 5 | 128.7 | |||
6 | 65.1 | 6 | 65.4 | 6 | 58.6 | |||
7 | 22.5 | 7 | 412 | 7 | 148.6 | |||
8 | 56.1 | 8 | 160.2 | 8 | 98.9 | |||
9 | 32.5 | 9 | 52.4 | 9 | 46.6 | |||
10 | 46.7 | 10 | 67.6 | 10 | 118.2 | |||
11 | 63.1 | 11 | 62.9 | 11 | 93.4 | |||
12 | 61.5 | 12 | 59.3 | 12 | 68.7 | |||
13 | 67.0 | 13 | 72.3 | 13 | 52.8 | |||
14 | 52.9 | 14 | 46.5 | 14 | 74.4 | |||
15 | 32.1 | 15 | 82.4 | 15 | 77.5 | |||
16 | 55.5 | 16 | 67.9 | 16 | 46.7 | |||
17 | 84.0 | 17 | 40.8 | 17 | 73.6 | |||
18 | 45.2 | 18 | 39.7 | 18 | 87.3 | |||
19 | 59.6 | 19 | 86.9 | 19 | 76.8 | |||
20 | 71.4 | 20 | 33.3 | 20 | 45.1 | |||
21 | 32.0 | 21 | 83.4 | 21 | 51.9 | |||
22 | 27.8 | 22 | 51.8 | 22 | 38.7 | |||
23 | 66.2 | 23 | 41.8 | 23 | 83.6 | |||
24 | 59.9 | 24 | 53.6 | 24 | 55.6 | |||
25 | 43.0 | 25 | 41.0 | 25 | 76.9 | |||
26 | 30.0 | 26 | 80.1 | 26 | 38.4 | |||
27 | 44.5 | 27 | 76.1 | 27 | 62.4 | |||
28 | 48.9 | 28 | 91.0 | 28 | 49.9 | |||
29 | 54.9 | 29 | 77.0 | 29 | 947 | |||
30 | 76.5 | 30 | 46.0 | 30 | 51.1 | |||
31 | 64.4 | 31 | 68.6 | 31 | 60.5 | |||
Population | Serial number | DNA μg/ml | Population | Serial number | DNA μg/ml | |||
MP | 1 | 47.3 | LP | 1 | 46.3 | |||
2 | 98.1 | 2 | 58.7 | |||||
3 | 45.5 | 3 | 40.0 | |||||
4 | 39.3 | 4 | 31.2 | |||||
5 | 151.0 | 5 | 47.3 | |||||
6 | 108.1 | 6 | 38.4 | |||||
7 | 73.8 | 7 | 29.9 | |||||
8 | 58.0 | 8 | 59.9 | |||||
9 | 71.0 | 9 | 56.8 | |||||
10 | 69.3 | 10 | 23.2 | |||||
11 | 38.6 | 11 | 57.1 | |||||
12 | 54.8 | 12 | 37.5 | |||||
13 | 94.7 | 13 | 77.7 | |||||
14 | 129.6 | 14 | 51.8 | |||||
15 | 58.8 | 15 | 53.4 | |||||
16 | 76.1 | 16 | 44.8 | |||||
17 | 85.7 | 17 | 40.9 | |||||
18 | 71.5 | 18 | 49.6 | |||||
19 | 69.9 | 19 | 63.2 | |||||
20 | 57.0 | 20 | 66.1 | |||||
21 | 60.8 | 21 | 57.2 | |||||
22 | 58.5 | 22 | 70.3 | |||||
23 | 76.8 | 23 | 55.0 | |||||
24 | 37.0 | 24 | 55.0 | |||||
25 | 67.0 | 25 | 68.0 | |||||
26 | 84.6 | 26 | 63.0 | |||||
27 | 89.0 | 27 | 31.0 | |||||
28 | 69.3 | 28 | 39.1 | |||||
29 | 67.9 | 29 | 45.8 | |||||
30 | 61.5 | 30 | 47.3 | |||||
31 | 67.0 | 31 | 66.5 |
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, L., Cao, B., Xie, S. et al. Genetic diversity of wild Camellia oleifera in northern China revealed by simple sequence repeat markers. Genet Resour Crop Evol (2023). https://doi.org/10.1007/s10722-023-01785-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-023-01785-4