Abstract
The Human Betaherpesviruses HHV-5 and HHV-6 are quite inimical in immunocompromised hosts individually. A co-infection of both has been surmised to be far more disastrous. This can be attributed to a synergetic effect of their combined pathologies. While there have been attempts to develop a vaccine against each virus, no efforts were made to contrive an effective prophylaxis for the highly detrimental co-infection. In this study, an ensemble of viral envelope glycoproteins from both the viruses was utilized to design a multi-epitope vaccine using immunoinformatics tools. A collection of bacterial protein toll-like receptor agonists (BPTAs) was screened to identify a highly immunogenic adjuvant for the vaccine construct. The constructed vaccine was analysed using an array of methodologies ranging from World population coverage analysis to Immune simulation, whose results indicate high vaccine efficacy and stability. Furthermore, codon optimization and in silico cloning analysis were performed to check for efficient expression in a bacterial system. Collectively, these findings demonstrate the potential of the constructed vaccine to elicit an immune response against HHV-5 and HHV-6, thus supporting the viability of in vitro and in vivo studies.
Similar content being viewed by others
Introduction
Human herpesvirus 5 (HHV-5), also known as the Human Cytomegalovirus (HCMV), belonging to the betaherpesvirus subfamily is the cause of one of the most prevalent diseases affecting immunocompromised hosts [1]. Belonging to the same subfamily, Human herpesvirus 6 (HHV-6) is an immunotropic virus that specifically attacks cells involved in both, humoral and cell-mediated immune responses [2]. Among the Human herpesviruses, HHV-5 HHV-6 co-infection is the one most frequently associated with post-immunosuppression complications [3].
It has been observed that HHV-5 and HHV-6 remain in a latent state after the primary infection until reactivation due to an immunosuppressive condition. Spreading through body fluids, HHV-5 has a global seroprevalence of about 94% [4]. Congenital infection may cause complications including neurological issues, hearing loss, visual impairment, and death. Among healthy adults, the primary infection may cause fatigue, fever, muscle aches, and rarely mononucleosis. The most common symptoms following primary infection in immunocompromised adults include pneumonia, vision loss, encephalitis, and digestive system issue [5]. HHV-6 is usually prevalent among infants, and manifests in the form of skin rashes known as roseola. Other symptoms include high fever, encephalitis, meningitis, liver dysfunction, or abnormalities such as granulocytopenia, wherein a very low white blood cell count is observed [6]. Co-infection of HHV-5 and HHV-6 mostly occurs due to their reactivation during immunosuppressive conditions such as chemotherapy or organ transplantation. Studies show that higher activation rates of HHV-5 can be observed in cases with preliminary HHV-6 infection [3]. The immunomodulatory and immunosuppressive effects of HHV-6 can induce a predisposition to HHV-5 infection or its reactivation. With their pathologies having a synergetic effect, it can be understood that a co-infection would have worse clinical implications than infection with either one of the viruses [3]. This synergetic co-infection has also been shown to increase the risk of fungal and bacterial infections [3, 7].
The current strategy to treat an HHV-5 infection is a 6-month prescription of the antiviral drugs Valganciclovir and Cidofovir [1]. This strategy is limited due to not being cost-effective and the safety concerns associated with drugs [1, 8]. For HHV-6, antiviral drugs Foscarnet and Valganciclovir or combinations of them are used [9]. This too is limited by their toxicity, antiviral resistance and drug-drug interactions [10]. Another major disadvantage is the fact that antiviral drugs only target lytic infections and not latent ones [11]. Vaccination, on the other hand, can lead to the complete eradication of the viruses on a geographical level. While vaccination does not prevent the reactivation of latent infections, it would significantly decrease the rates of primary infections [12]. Various approaches for HHV-5 vaccine development were considered in previous studies, ranging from live attenuated vaccines to DNA, RNA or protein subunit-based vaccines [11]. While live attenuated vaccines such as the Towne attenuated strain [12] or the Towne-Toledo recombinant strain [13] showed significant immunogenicities, they are limited by their inherent risk of causing the disease they are supposed to provide immunity from [14]. DNA vaccines based on the pp65 and gB [15, 16] induced immune responses in 47.5–68% of the seronegative subjects and an RNA vaccine based on gB and pp65/IE1 fusion protein [17] showed antibody and T-cell responses in almost 97% of the subjects after the second dose. A recombinant protein vaccine derived from the envelope gB showed only 50% efficacy against a primary HHV-5 infection [18]. Though vaccine development for HHV-6 did not receive the same scrutiny that HHV-5 immunization endeavours did, efforts were put in to identify potential vaccine candidates based on viral protein complexes. A tetramer complex of envelope glycoproteins gH, gL, gQ1 and gQ2 was observed to induce immunization in mice against HHV-6 [19]. This tetramer targets the human CD134 host receptor, which is expressed on activated T lymphocytes. This interaction determines the HHV-6B cell tropism and is necessary for HHV-6B entry into the vulnerable cells.
Till date, there have been no attempts to develop a vaccine against a co-infection of HHV-5 and HHV-6. In this scenario, immunoinformatics tools were used to construct a multi-epitope vaccine candidate in order to provide immunity against a synergetic co-infection of the two betaherpesviruses. In essence, the present computational vaccine design study with aid of unique glycoprotein is expressed on the virus envelope of human herpesvirus is indeed an important supplement in the scientific literature of glycoprotein based therapeutics. The strain such as AD169 for HHV-5 and U-1102 for HHV-6 were chosen based on their quality, completeness and validity. We hope that present study employing immunoinformatics pipeline would provide a safer platform for vaccine development and provide clue for the experimental biologist working in this field.
Methodology
Amino acid sequence retrieval
In this study, an ensemble of computational tools was adopted to predict ideal vaccine candidates for the HHV-5 and HHV-6 using viral envelope glycoproteins. For the epitope prediction, a total of 9 envelope glycoproteins from HHV-5 and 7 envelope glycoproteins from HHV-6 were chosen. Each of these glycoproteins was selected due to their roles in viral attachment to the host cells and the fusion of viral and host plasma membranes [20,21,22,23, 25, 26]. The protein sequences were retrieved from the UniProt database (https://www.uniprot.org/uniprot/P06473). Of note, the strains AD169 (HHV-5) and U-1102 (HHV-6) envelope glycoprotein sequences were chosen because they were longest and experimentally verified sequence with highest annotation scores.
Linear B-cell epitope prediction
B-cell epitope prediction was done using the ABCpred tool (http://crdd.osdd.net/raghava/abcpred/). The tool with a prediction accuracy of 65.93% utilizes a Recurrent Neural Network trained using a dataset consisting of 700 B-cell epitopes and 700 non-B-cell epitopes retrieved from the SwissProt database [28]. For the current study, a score threshold of 0.7 and an epitope length window of 10 were chosen for the predicted epitopes.
T-cell epitope prediction
Cytotoxic T-cell epitope prediction
Cytotoxic T lymphocyte (CTL) epitope prediction from the viral glycoprotein sequences was done using the NetMHCpan 4.1 tool (https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.1). NetMHCpan 4.1 employs an Artificial Neural Networks (ANNs) to predict the peptide binding to any MHC molecule whose sequence is known. For the current analysis, 12 MHC I Allele subgroup belonging to the 2 major HLA (A and B) super types were selected. The predicted peptide length set to 9mer (Table S1). An epitope identification threshold of 0.5% was set for strong binders and 2% for weak binders [29].
Helper T-cell epitope prediction
Epitopes specific to Helper T lymphocytes (HTL) were predicted from the viral protein sequences using NetMHCIIpan 4.0 tool (https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.0). A total of 27 MHCII alleles subgroup belonging to the 3 HLA (DR, DP and DQ) super types were considered (Table S1), as the binding efficacy of HTL epitopes to HLA-DR is a key factor in the immunogenicity of T-cell epitopes [30]. 15mer epitopes were predicted with the selection threshold at 1% for strong binders and 5% for weak binders [29].
Epitope characterization
Toxicity evaluation
ToxinPred tool (http://crdd.osdd.net/raghava/toxinpred/) which utilizes a Support Vector Machine trained using a dataset of toxic and non-toxic peptides was used to evaluate the toxicity of each predicted BCL, CTL and HTL epitopes along with important physicochemical properties like charge, isoelectric point (pI), hydrophobicity, etc. [31]. The understanding of vaccine’s physicochemical qualities is of useful to gain insight into the vaccine construct interaction with the environment. For instance, theoretical pI of a protein would be helpful in order to choose and improve the protein purification techniques.
Allergenicity prediction
The allergenicity of the epitopes was checked using the AllerTop 2.0 server (https://www.ddg-pharmfac.net/AllerTOP/) which uses a k-nearest neighbor (kNN) classifier trained on 2427 allergens and an equal number of non-allergens [32].
Antigenicity prediction
Antigenicity gives a measure of an antigen’s abilty to bind to B-cell and T-cell receptors, thereby eliciting an immune response. To identify probable antigenic epitopes, Vaxijen 2.0 server was used with a threshold of 0.4 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). Vaxijen utilizes alignment independent prediction (89% accuracy) based solely on the physicochemical properties of the proteins [33].
CTL Immunogenicity prediction
Immunogenicity evaluation for the predicted epitopes was done using IEDB class-I immunogenicity tool (http://tools.iedb.org/immunogenicity/) run with default parameters. The tool utilizes amino acid properties and their position within the peptide to make the immunogenicity prediction for a peptide-MHC complex [34].
Vaccine construction
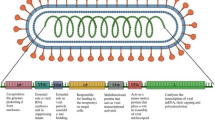
To elicit an immune response, protein subunit vaccines must consist of an antigenic component from the pathogen. To accomplish this, the screened CTL, HTL and BCL epitopes along with an adjuvant were combined in a sequence using appropriate linkers. An adjuvant is any component that induces humoral or cellular immune response against an antigen [35]. Bacterial proteins with potential TLR agonist activity were considered as the adjuvants for the vaccine candidate [36]. The vaccine construct sequence begins with the adjuvant followed by BCL epitopes, with the first BCL epitoped joined to the adjuvant via an EAAAK linker. The BCL epitopes were attached to each other using KK linkers, and AAY linkers were used to attach CTL epitopes to the sequence, followed by HTL epitopes attached using GPGPG linkers.
Population coverage
Due to ethnic and geographic variations in the Human leukocyte Antigen (HLA) alleles, estimating the percentage of individuals covered by the respective HLA alleles is essential for ensuring vaccine efficacy. To determine the global population coverage, the IEDB population coverage analysis tool (http://tools.iedb.org/population/) was utilized with HLA class I and class II combined [37]. The analysis was carried out for T-cell epitopes along with their corresponding HLA alleles.
Physicochemical feature prediction
ProtParam tool (https://web.expasy.org/protparam/) was used to predict the physicochemical properties including atomic composition, amino acid composition, aliphatic index, molecular weight (kDa), extinction coefficient, theoretical pI, grand average of hydrophobicity (GRAVY), estimated half-life, and instability index of the constructed vaccine [38]. For performing a comparative evaluation of the physicochemical properties of the construct, postive controls C1 (Human Cytomegalovirus peptide vaccine) [39], C2 (typhoidal Salmonella serovars) [40], C3 (SARS-Cov-2 glycoprotein) [41] were chosen.
Secondary structure prediction
The secondary structure prediction for the vaccine construct was done using PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/) which utilizes PSI-BLAST (Position-Specific Iterated-BLAST) to predict the secondary structure and an artificial neural network to predict the alpha helices, beta sheets and coils of the construct [42].
Tertiary structure prediction and its validation
Robetta server (https://robetta.bakerlab.org/) was utilized for predicting the tertiary structure of the constructed vaccine [43]. Validation for the predicted structure was done using PROCHECK, Verify 3D and ERRAT tools from the SAVES 6.0 server (https://saves.mbi.ucla.edu/). PROCHECK tool enables the construction of Ramachandran plots to evaluate the generated structure models [44]. The overall quality factor was determined using ERRAT [45]. Verify 3D utilizes DSSP- a hydrogen bond estimation algorithm for further validation that works by calculating the most likely secondary structure assignment for a given 3D structure [46].
Conformational B-cell epitope prediction
Conformational/Discontinuous B-cell epitopes arise due to conformational changes brought about by the physicochemical folding in the tertiary structure. Conformational epitope prediction was done using the ElliPro tool (http://tools.iedb.org/ellipro/) run with default parameters. Based on the geometrical properties of the construct, Ellipro utilizes Thornton’s method along with a residue clustering algorithm to make the predictions, and a Protrusion Index (PI) score is assigned to each predicted epitope [47].
Vaccine candidate-Immune receptor molecular docking analysis
In silico evaluation of the binding correlation between the vaccine candidate and an immune receptor was performed using the ClusPro server (https://cluspro.bu.edu/login.php) [32]. TLR4 was the receptor chosen, its PDB structure was retrieved from the PDB database and prepared for docking using PyMol visualization software. The interaction between the vaccine and the TLR4 was analysed to gain insight into the binding pattern.
Immune simulation
Immune simulation was performed for the vaccine candidate using the C-ImmSim server (https://kraken.iac.rm.cnr.it/C-IMMSIM/). C-ImmSim is an agent-based model implementation, which utilizes position-specific scoring matrices (PSSM) and machine learning models for epitope and immune response predictions defining both humoral and cellular responses of the immune system [48]. Three injections were given at 1, 84 and 168 simulation time steps (each 4 weeks apart), and the simulation was run for a total of 1050 time steps. Each injection contained 1000 vaccine proteins with no LPS.
Codon optimization and in-silico cloning
The amino acid sequence of the vaccine construct was converted to its nucleotide sequence and the resulting codons were adapted for efficient expression in E.coli K12, using the Java Codon adaptation tool (Jcat) (http://www.jcat.de/). Using SnapGene the optimized codon sequence was cloned into a common expression vector pET-28a ( +).
Result and discussion
Viral protein selection
The sequence of the viral envelope glycoproteins gB, gH, gL, gM, gN, gO, UL128, UL130, UL131A of HHV-5 (strain AD169) and gB, gH, gL, gM, gN, gQ1, gQ2 of HHV-6 (strain U-1102) were retrieved from the Uniprot database. Glycoprotein B (gB) present in both the viruses, plays a major role in virus entry into the host cell and intercellular spread [20, 21]. HHV-5 glycoproteins gH and gL form two alternative complexes, a trimer of gH/gL/gO which binds to the PDGFR-α on fibroblast surfaces [22] and a pentamer of gH/gL/gO/UL128/UL130/UL131A which is involved in the virion internalization into monocytes [23]. In HHV-6, a tetramer complex of gH/gL/gQ1/gQ2 along with gB is required for the fusion of viral and host cell membranes [21]. In both HHV-5 and HHV-6, gM and gN form a complex that is essential for membrane fusion and intracellular transport [24, 25]. The gM/gN complex has also been observed to induce the production of HHV-5 neutralizing antibodies in vitro [26] while gN alone has implications in protecting the virus from antibodies [27].
Linear B-cell epitope prediction
B-cell epitopes are parts of the antigen that bind to the antibodies, allowing B-cells to neutralize pathogens contributing to humoral immunity [49]. Using the ABCPred tool, epitope prediction was carried out with default threshold and epitope length set to 10mer, resulting in predicted 577 BCL epitopes. Out of these, 166 epitopes with a prediction score greater than 0.7 were selected. These were subjected to screening by checking toxicity and allergenicity via ToxinPred and AllerTop, respectively. This yielded 49 non-toxic and non-allergenic epitopes. The final 7 BCL epitopes were selected based on antigenicity scores predicted using Vaxijen. These include RHRDVQHGRR, RRRDVGDVKS, LTFSDRETLF, VKDQWHSRGS, ALLLKFNNLG, VTALSFRLVA and VKSTPPPEDK. The details of the BCL epitopes are illustrated in Table 1.
T-cell epitope prediction
Cytotoxic T-cell epitope mapping
Cytotoxic T lymphocytes (CTLs) play a vital role in cell-mediated immunity destroying infected cells via antigen recognition. T-cell epitopes are the peptides that activate CTLs by binding to MHC-I proteins, thereby enabling recognition. NetMHCpan 4.1 server was used to predict CTL epitopes from the viral protein sequence, yielding 5505 CTL epitopes. Out of these 1241 epitopes were classified as strong binders. Here 286 epitopes with %Rank-EL less than 0.5 were selected for further analysis. These epitopes were screened based on their predicted toxicity, allergenicity, and antigenicity scores yielding 91 epitopes. Further screening was performed, to select epitopes with antigenicity scores greater than 1.0 and immunogenicity scores greater than 0.30, yielding the following 7 epitopes DVIDVQYRF, ALSFRLVAL, QLVDLTLNY, YSNIGFLLY, ALSFINVTV, LLRHHFHCL and HMFFTNLTF for further analysis (Table 2).
Helper T-cell epitope mapping
Helper T lymphocytes (HTLs) play an indispensable role in giving rise to the adaptive immune response, by utilizing signalling molecules to activate B-cells, cytotoxic T-cells and macrophages [50]. HTL epitopes were predicted using the NetMHCIIpan 4.0 server, yielding 6329 epitopes. Out of these 689 were classified as strong binders. A total of 327 epitopes with a %Rank-EL less than 1.0 were selected for further characterization. 67 Non-toxic and non-allergenic epitopes were further screened, to select epitopes with antigenicity scores greater than 1.0, yielding the following 7 epitopes LDFNYLDLSALLRNS, YFEINDLKAVNLSAN, SFFAFQKIHPNLKGT, IVHFSYSTKNTGPMP, ALSFRLVALGAFAYC, VEALLLKFNNLGIQT and IDPLENTDFRVLELY for further analysis (Table 3).
Vaccine construction
A total of 21 epitopes (7 BCL, 7 CTL and 7 HTL) were used to make the final vaccine construct (Fig. 1). Appropriate linkers were used to connect the epitopes sequentially. For instance, an EAAAK linker is used to connect the adjuvant and BCL epitopes, a KK linker to connect BCL epitopes, an AAY linker to connect CTL epitopes and finally, a GPGPG linker to connect HTL epitopes. Out of the candidate bacterial protein adjuvants considered, CobT protein from Mycobacterium paratuberculosis, imparted the highest immunogenicity and hence it is chosen as the adjuvant for the final vaccine construct (Table 4).
Population coverage analysis
The frequency of different HLA alleles varies with ethnicities and geographic regions. This necessitates determining the percentage of the global population covered by HLA alleles of the predicted epitopes. The results predict 99.95% global population coverage for the constructed vaccine (Table S2, S3 and Figure S1).
Physicochemical features and secondary structure prediction
ProtParam tool was used to determine the physicochemical properties of the final vaccine construct and the positive controls C1, C2 and C3. The predicted features for the final construct (Vaccine + CobT) include a length of 660aa, a molecular weight of 69.52 kDa indicating good antigenicity and a theoretical PI of 8.96 (basic behaviour). The instability index of the protein was calculated to be 23.70, classifying the protein as stable. The half-life of the protein was estimated to be 30 h in mammalian reticulocytes (in vitro), > 20 h in yeast (in vivo), and > 10 h in Escherichia coli (in vivo). An aliphatic index of 92.12 indicates moderate stability of the protein in vivo. The grand average of hydropathicity (GRAVY) score of -0.073 indicates a slightly hydrophilic nature of the protein. These properties are similar to those predicted for the controls C1, C2, and C3 (Table 4). The secondary structure prediction for the vaccine was performed using the PSIPRED tool, which indicated the construct to be 16.89% alpha-helices, 37.08% strands and 46.01% coils (Fig. 2).
Tertiary structure prediction and its validation
The tertiary structure of a protein provides different structural insights and is predicted via homology modelling. ROBETTA server was used for the 3D structure prediction, followed by its validation PROCHECK, ERRAT and Verify 3D tools from SAVES 6.0 server. Of the 5 models predicted by ROBETTA, Model-2 was the ideal choice (Figure S2). The Ramachandran plots from PROCHECK showed 92.40% of residues in the most favoured regions and only 0.40% of residues in disallowed regions. ERRAT predicted an overall quality factor of 97.8528 and Verify 3D demonstrated the overall structure accuracy to be 84.85% (Table S4). The 3D structure of the vaccine construct was visualized using PyMol (Fig. 3).
Conformational B-cell epitope prediction
Conformational/Discontinuous epitopes mimic the properties of a linear epitope and can be used as a replacement for antibody production. Potential conformational epitopes were predicted using the ElliPro tool, yielding 5 conformational B-cell epitopes (Table S5). The predicted epitopes were ranked based on their Protrusion Index (PI) scores. The 3D structure of the top-ranking epitope consisting of 76 residues was visualized using jmol – a molecular viewer integrated within ElliPro (Fig. 4).
Vaccine construct-TLR4 docking studies
The envelope glycoproteins of HHV5 and HHV6 have been shown to effectively be recognized by TLR4 [51]. To determine this interaction, protein–protein docking between the vaccine construct and TLR4 (PDB ID: 3FXI) was performed using ClusPro 2.0 server. From among the resulting models, the one with the largest cluster (52) and least energy (-1084.8) was selected. The larger the cluster size, the greater the certainty of the cluster’s centre representing a putative model of the complex. Figure 5 illustrates the 3D structure of the docked complex. Further, the molecular interactions was visualized using the PDBSum. From Fig. 6, it can be observed that chains A and B from TLR4 interacted with the designed vaccine. There were 2 hydrogen bonds between chain B and the vaccine and a total of 13 hydrogen bonds between the residues of chain A and the vaccine. These interactions suggest that our proposed vaccine has the ability to be recognized by the T cell receptors.
Immune Simulation
The effectiveness of the vaccine construct to elicit an appreciable response from the adaptive immune system was determined using the C-Immsim server. Graphs of antibody, cytokine and lymphocyte concentrations were generated (Fig. 7). The antibody titre (IgG + IgM) rises after every dose and is the highest after the third dose. A simultaneous decrease in the antigen concentration was also observed after successive injections. Significant spikes in the B-cell and T-cell concentrations were also evident. Higher IgM concentrations ensure B-cell activation and the subsequent production of antibodies required for the secondary and tertiary immune responses. The vaccine has also been observed to induce high levels of IL-2 which has an immunoregulatory role and INF-g which is involved in APC activation.
Codon optimization and in-silico cloning
The Java Codon adaptation tool (JCat) yielded an improved codon sequence for the vaccine construct sequence, for ensuring efficacious expression in E.coli K12. The improved sequence showed a GC content of 56.46% and a Codon Adaptation Index (CAI) of 0.9975 signifying sequence stability and optimized expression in the host, respectively. The improved codon sequence was cloned between HpaI (1629) and SmaI (4300) of pET-28a ( +) which is a well-known expression vector resulting in a cloned vector of 4331 bp (Fig. 8).
Conclusion
In the current investigation, a multi-epitope vaccine construct was generated using highly antigenic and immunogenic B-cell and T-cell epitopes predicted from the envelope glycoproteins of HHV-5 and HHV-6. Linkers such as KK, AAY and GPGPG were utilized to join the epitopes and an adjuvant was attached to boost its immunogenicity. The secondary and tertiary structures of the vaccine construct were predicted along with their physicochemical properties. The helix-rich nature of the construct along with metrics such as the instability index and aliphatic index reflects the stability of the constructed vaccine. Taking into account the global polymorphism of the MHC class I and class II molecules, a world population coverage analysis was conducted, and the results indicated that our proposed vaccine would induce an immunological response in 99.95% of the world's population. Similarly, molecular docking with the immune receptor TLR4 was performed and the results signifies the high binding affinity and stability of the vaccine-receptor complex. The immune simulation study showed a significant production of antibodies and cytokines after injecting vaccine. Finally, the nucleotide sequence for the vaccine construct was determined and the codons were adapted to be efficiently expressed in a bacterial host. In conclusion, the constructed vaccine has the potential to be translated into clinical practice to overcome the burden of HHV-5 and HHV-6 co-infection. The experimental studies on vaccine's efficacy in animal models is an interesting future direction and of very much importance to validate this findings.
Data availability
Provided as Required.
Code availability
Not Applicable.
Abbreviations
- HHV-5:
-
Human Herpesvirus 5
- HHV-6:
-
Human Herpesvirus 6
- HCMV:
-
Human Cytomegalovirus
- PDGFR:
-
Platelet-derived growth factor receptor
- MHC:
-
Major Histocompatibility Complex
- HLA:
-
Human Leukocyte Antigen
- IEDB:
-
Immune Epitope Database
- PDB:
-
Protein Data Bank
- TLR4:
-
Toll-like Receptor 4
References
Griffiths, P.D.: Burden of disease associated with human cytomegalovirus and prospects for elimination by universal immunisation. Lancet. Infect. Dis. 10, 790–798 (2012). https://doi.org/10.1016/S1473-3099(12)70197-4
Lusso, P.: HHV-6 and the immune system: mechanisms of immunomodulation and viral escape. J. Clin. Virol. S1, S4-S10 (2006). https://doi.org/10.1016/S1386-6532(06)70004-X
Handous, I., Achour, B., Marzouk, M., Rouis, S., Hazgui, O., Brini, I., Khelif, A., Hannachi, N., Boukadida, J.: Co-infections of human herpesviruses (CMV, HHV-6, HHV-7 and EBV) in non-transplant acute leukemia patients undergoing chemotherapy. Virol. J. 17(1), 37 (2020). https://doi.org/10.1186/s12985-020-01302-4
Ssentongo, P., Hehnly, C., Birungi, P., Roach, M.A., Spady, J., Fronterre, C., Wang, M., Murray-Kolb, L.E., Al-Shaar, L., Chinchilli, V.M., Broach, J.R., Ericson, J.E., Schiff, S.J.: Congenital Cytomegalovirus Infection Burden and Epidemiologic Risk Factors in Countries With Universal Screening: A Systematic Review and Meta-analysis. JAMA Netw Open. 4(8), e2120736 (2021). https://doi.org/10.1001/jamanetworkopen.2021.20736
Manicklal, S., Emery, V.C., Lazzarotto, T., Boppana, S.B., Gupta, R.K.: The "silent" global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 26(1), 86–102 (2013). https://doi.org/10.1128/CMR.00062-12
Lusso, P.: Human herpesvirus 6 (HHV-6). Antiviral. Res. 31(1–2), 1–21 (1996). https://doi.org/10.1016/0166-3542(96)00949-7
Reymen, D., Naesens, L., Balzarini, J., Holý, A., Dvoráková, H., Clercq, E De.: Antiviral activity of selected acyclic nucleoside analogues against human herpesvirus 6. Antiviral. Res. 28(4), 343–357 (1995). https://doi.org/10.1016/0166-3542(95)00058-5
Seelbinder, B., Wallstabe, J., Marischen, L., Weiss, E., Wurster, S., Page, L., Löffler, C., Bussemer, L., Schmitt, A.L., Wolf, T., Linde, J., Cicin-Sain, L., Becker, J., Kalinke, U., Vogel, J., Panagiotou, G., Einsele, H., Westermann, A.J., Schäuble, S., Loeffler, J.: Triple RNA-Seq Reveals Synergy in a Human Virus-Fungus Co-infection Model. Cell Rep. 33(7), 108389 (2020). https://doi.org/10.1016/j.celrep.2020.108389
Gardner, T.J., Tortorella, D.: Virion Glycoprotein-Mediated Immune Evasion by Human Cytomegalovirus: a Sticky Virus Makes a Slick Getaway. Microbiol. Mol. Biol. Rev. 80(3), 663–677 (2016). https://doi.org/10.1128/MMBR.00018-16
Ogata, M., Satou, T., Kadota, J., Saito, N., Yoshida, T., Okumura, H., Ueki, T., Nagafuji, K., Kako, S., Uoshima, N., Tsudo, M., Itamura, H., Fukuda, T.: Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: a multicenter, prospective study. Clin. Infect. Dis. 57(5), 671–681 (2013). https://doi.org/10.1093/cid/cit358
Gugliesi, F., Coscia, A., Griffante, G., Galitska, G., Pasquero, S., Albano, C., Biolatti, M.: Where do we Stand after Decades of Studying Human Cytomegalovirus? Microorganisms. 8(5), 685 (2020). https://doi.org/10.3390/microorganisms8050685
Plotkin, S.A., Furukawa, T., Zygraich, N., Huygelen, C.: Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 2(3), 521–527 (1975). https://doi.org/10.1128/iai.12.3.521-527.1975
Adler, S.P., Manganello, A.M., Lee, R., McVoy, M.A., Nixon, D.E., Plotkin, S., Mocarski, E., Cox, J.H., Fast, P.E., Nesterenko, P.A., Murray, S.E., Hill, A.B., Kemble, G.: A Phase 1 Study of 4 Live, Recombinant Human Cytomegalovirus Towne/Toledo Chimera Vaccines in Cytomegalovirus-Seronegative Men. J. Infect. Dis. 214(9), 1341–1348 (2016). https://doi.org/10.1093/infdis/jiw365
Minor, P.D.: Live attenuated vaccines: Historical successes and current challenges. Virology. 479(480), 379–392 (2015). https://doi.org/10.1016/j.virol.2015.03.032
Wloch, M. K.: Safety and immunogenicity of a bivalent cmv dna vaccine in healthy adult subjects. J. Infect. Dis. 197, 1634–1642 (2010). https://doi.org/10.1086/588385
Smith, L.R., Wloch, M.K., Chaplin, J.A., Gerber, M., Rolland, A.P.: Clinical Development of a Cytomegalovirus DNA Vaccine: From Product Concept to Pivotal Phase 3 Trial. Vaccines (Basel). 1(4), 398–414 (2013). https://doi.org/10.3390/vaccines1040398
Bernstein, D.I., Reap, E.A., Katen, K., Watson, A., Smith, K., Norberg, P., Olmsted, R.A., Hoeper, A., Morris, J., Negri, S., Maughan, M.F., Chulay, J.D.: Randomized, double-blind, Phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine. 28(2), 484–493 (2009). https://doi.org/10.1016/j.vaccine.2009.09.135
Pass, R.F., Zhang, C., Evans, A., Simpson, T., Andrews, W., Huang, M.L., Corey, L., Hill, J., Davis, E., Flanigan, C., Cloud, G.: Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 360(12), 1191–1199 (2009). https://doi.org/10.1056/NEJMoa0804749
Wang, B., Hara, K., Kawabata, A., Nishimura, M., Wakata, A., Tjan, L.H., Poetranto, A.L., Yamamoto, C., Haseda, Y., Aoshi, T., Munakata, L., Suzuki, R., Komatsu, M., Tsukamoto, R., Itoh, T., Nishigori, C., Saito, Y., Matozaki, T., Mori, Y.: Tetrameric glycoprotein complex gH/gL/gQ1/gQ2 is a promising vaccine candidate for human herpesvirus 6B. PLoS. Pathog. 16(7), e1008609 (2020). https://doi.org/10.1371/journal.ppat.1008609
Isaacson, M.K., Compton, T.: Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J. Virol. 83(8), 3891–3903 (2009). https://doi.org/10.1128/JVI.01251-08
Tanaka, Y., Suenaga, T., Matsumoto, M., Seya, T., Arase, H.: Herpesvirus 6 glycoproteins B (gB), gH, gL, and gQ are necessary and sufficient for cell-to-cell fusion. J. Virol. 87(19), 10900–10903 (2013). https://doi.org/10.1128/JVI.01427-13
Wu, Y., Prager, A., Boos, S., Resch, M., Brizic, I., Mach, M., Wildner, S., Scrivano, L., Adler, B.: Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS. Pathog. 13(4), e1006281 (2017). https://doi.org/10.1371/journal.ppat.1006281
Nogalski, M.T., Chan, G.C., Stevenson, E.V., Collins-McMillen, D.K., Yurochko, A.D.: The HCMV gH/gL/UL128–131 complex triggers the specific cellular activation required for efficient viral internalization into target monocytes. PLoS. Pathog. 9(7), e1003463 (2013). https://doi.org/10.1371/journal.ppat.1003463
Mach, M., Kropff, B., Kryzaniak, M., Britt, W.: Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79(4), 2160–2170 (2005). https://doi.org/10.1128/JVI.79.4.2160-2170.2005
Kawabata, A., Serada, S., Naka, T., Mori, Y.: Human herpesvirus 6 gM/gN complex interacts with v-SNARE in infected cells. J. Gen. Virol. 95(12), 2769–2777 (2014). https://doi.org/10.1099/vir.0.069336-0
Shimamura, M., Mach, M., Britt, W.J.: Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J. Virol. 80(9), 4591–4600 (2006). https://doi.org/10.1128/JVI.80.9.4591-4600.2006
Kropff, B., Burkhardt, C., Schott, J., Nentwich, J., Fisch, T., Britt, W., Mach, M.: Glycoprotein N of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS. Pathog. 8(10), e1002999 (2012). https://doi.org/10.1371/journal.ppat.1002999
Saha, S., Raghava, G.P.: Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 65(1), 40–48 (2006). https://doi.org/10.1002/prot.21078
Reynisson, B., Alvarez, B., Paul, S., Peters, B., Nielsen, M.: NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic. Acids. Res. 48(1), 449–454 (2020). https://doi.org/10.1093/nar/gkaa379
Lazarski, C.A., Chaves, F.A., Jenks, S.A., Wu, S., Richards, K.A., Weaver, J.M., Sant, A.J.: The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 23(1), 29–40 (2005). https://doi.org/10.1016/j.immuni.2005.05.009
Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R.: Open Source Drug Discovery Consortium, Raghava GP. In silico approach for predicting toxicity of peptides and proteins. PLoS. One. 8(9), e73957 (2013). https://doi.org/10.1371/journal.pone.0073957
Kozakov, D., Hall, D.R., Xia, B., Porter, K.A., Padhorny, D., Yueh, C., Beglov, D., Vajda, S.: The ClusPro web server for protein-protein docking. Nat. Protoc. 12(2), 255–278 (2017). https://doi.org/10.1038/nprot.2016.169
Doytchinova, I.A., Flower, D.R.: VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 8(4), (2007). https://doi.org/10.1186/1471-2105-8-4
Calis, J.J., Maybeno, M., Greenbaum, J.A., Weiskopf, D., De Silva, A.D., Sette, A., Keşmir, C., Peters, B.: Properties of MHC class I presented peptides that enhance immunogenicity. PLoS. Comput. Biol. 9(10), e1003266 (2013). https://doi.org/10.1371/journal.pcbi.1003266
Lima, K.M., dos Santos, S.A., Rodrigues, J.M., Silva, C.L.: Vaccine adjuvant: it makes the difference. Vaccine. 22(19), 2374–2379 (2004). https://doi.org/10.1016/j.vaccine.2003.12.030
Kumar, S., Sunagar, R., Gosselin, E.: Bacterial Protein Toll-Like-Receptor Agonists: A Novel Perspective on Vaccine Adjuvants. Front. Immunol. 10, 1144 (2019). https://doi.org/10.3389/fimmu.2019.01144.
Bui, H. H., Sidney, J., Dinh, K., Southwood, S., Newman, M. J., Sette, A.: Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 7(1), 153 (2006). https://doi.org/10.1186/1471-2105-7-153.
Gasteiger, E.: The Proteomics Protocols Handbook. Proteomics Protoc. Handb. (2005).
Akhtar, N., Joshi, A., Singh, J., Kaushik, V.: Design of a novel and potent multivalent epitope based human cytomegalovirus peptide vaccine: an immunoinformatics approach. J. Mol. Liq. 335, 116586 (2021)
Mahapatra, S.R., Dey, J., Kushwaha, G.S., Puhan, P., Mohakud, N.K., Panda, S.K., Lata, S., Misra, N., Suar, M.: Immunoinformatic approach employing modeling and simulation to design a novel vaccine construct targeting MDR efflux pumps to confer wide protection against typhoidal Salmonella serovars. J. Biomol. Struct. Dyn. 31, 1–13 (2021). https://doi.org/10.1080/07391102.2021.1964600
Rahman, N., Al,i F., Basharat, Z., Shehroz, M., Khan, M. K., Jeandet, P., Nepovimova, E., Kuca, K., Khan H.: Vaccine Design from the Ensemble of Surface Glycoprotein Epitopes of SARS-CoV-2: An Immunoinformatics Approach. Vaccines (Basel). 8(3), 423 (2020). https://doi.org/10.3390/vaccines8030423
Jones, D. T.: Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292(2), 195–202 (1999). https://doi.org/10.1006/jmbi.1999.3091
Kim, D. E., Chivian, D., Baker, D.: Protein structure prediction and analysis using the Robetta server. Nucleic. Acids. Res. 32, 526–531 (2004). https://doi.org/10.1093/nar/gkh468
Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M.: PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26(2), 283–291 (1993)
Colovos, C., Yeates, T. O.: Verification of protein structures: patterns of nonbonded atomic interactions. Protein. Sci. 2(9), 1511–1519 (1993). https://doi.org/10.1002/pro.5560020916
Eisenberg, D., Lüthy, R., Bowie, J. U.: VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277, 396–404 (1997). https://doi.org/10.1016/s0076-6879(97)77022-8
Ponomarenko, J., Bui, H. H., Li, W., Fusseder, N., Bourne, P. E., Sette, A., Peters, B.: ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 9, 514 (2008). https://doi.org/10.1186/1471-2105-9-514
Rapin, N., Lund, O., Castiglione, F.: Immune system simulation online. Bioinformatics. 27(14), 2013–2014 (2011). https://doi.org/10.1093/bioinformatics/btr335
LeBien, T. W., Tedder, T. F.: B lymphocytes: how they develop and function. Blood. 112(5), 1570–1580 (2008). https://doi.org/10.1182/blood-2008-02-078071
Saravia, J., Chapman, N. M., Chi, H.: Helper T cell differentiation. Cell. Mol. Immunol. 16(7), 634–643 (2019). https://doi.org/10.1038/s41423-019-0220-6
Zheng, W., Xu, Q., Zhang, Y., E. X., Gao, W., Zhang, M., Zhai, W., Rajkumar, R. S., Liu, Z.: Toll-like receptor-mediated innate immunity against herpesviridae infection: a current perspective on viral infection signaling pathways. Virol. J. 17(1), 192 (2020). https://doi.org/10.1186/s12985-020-01463-2
Acknowledgements
The authors gratefully acknowledge Vellore Institute of Technology, Vellore for its support in carrying out this research work.
Author information
Authors and Affiliations
Contributions
R.K. designed the computational framework and analysed the data. B.G. and H.K. performed the computational work, prepared tables, and figures. B.G., H.K., S.M., and S.V authors contributed to the writing of the manuscript. R.K. supervised the entire study. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gandhamaneni, B.S., Krishnamoorthy, H.R., Veerappapillai, S. et al. Envelope Glycoprotein based multi-epitope vaccine against a co-infection of Human Herpesvirus 5 and Human Herpesvirus 6 using in silico strategies. Glycoconj J 39, 711–724 (2022). https://doi.org/10.1007/s10719-022-10083-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-022-10083-7