Abstract
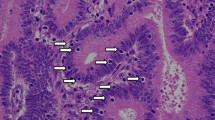
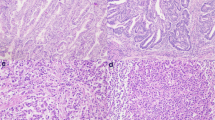
The widespread use of tumor DNA mismatch repair (MMR) protein immunohistochemistry in gastrointestinal tract (GIT) carcinomas has unveiled cases where the MMR protein status differs between synchronous/metachronous tumors from the same patients. This study aims at examining the frequency, patterns and molecular etiologies of such inter-tumoral MMR discordances. We analyzed a cohort of 2159 colorectal cancer (CRC) patients collected over a 5-year period and found that 1.3% of the patients (27/2159) had ≥ 2 primary CRCs, and 25.9% of the patients with ≥ 2 primary CRCs (7/27) exhibited inter-tumoral MMR discordance. We then combined the seven MMR-discordant CRC patients with three additional MMR-discordant GIT carcinoma patients and evaluated their discordant patterns and associated molecular abnormalities. The 10 patients consisted of 3 patients with Lynch syndrome (LS), 1 with polymerase proofreading-associated polyposis (PAPP), 1 with familial adenomatous polyposis (FAP), and 5 deemed to have no cancer disposing hereditary syndromes. Their MMR discordances were associated with the following etiologies: (1) PMS2-LS manifesting PMS2-deficient cancer at an old age when a co-incidental sporadic MMR-proficient cancer also occurred; (2) microsatellite instability-driven secondary somatic MSH6-inactivation occurring in only one—and not all—PMS2-LS associated MMR-deficient carcinomas; (3) “compound LS” with germline mutations in two MMR genes manifesting different tumors with deficiencies in different MMR proteins; (4) PAPP or FAP syndrome-associated MMR-proficient cancer co-occurring metachronously with a somatic MMR-deficient cancer; and (5) non-syndromic patients with sporadic MMR-proficient cancers co-occurring synchronously/metachronously with sporadic MMR-deficient cancers. Our study thus suggests that inter-tumoral MMR discordance is not uncommon among patients with multiple primary GIT carcinomas (25.9% in patients with ≥ 2 CRCs), and may be associated with widely varied molecular etiologies. Awareness of these patterns is essential in ensuring the most effective strategies in both LS detection and treatment decision-making. When selecting patients for immunotherapy, MMR testing should be performed on the tumor or tumors that are being treated.





Similar content being viewed by others
References
Boland PM, Yurgelun MB, Boland CR (2018) Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J Clin 68:217–231
Samowitz WS (2015) Evaluation of colorectal cancers for Lynch syndrome: practical molecular diagnostics for surgical pathologists. Mod Pathol 28(Suppl 1):S109–S113
Rybak C, Hall MJ (2011) Interpretation of genetic testing for Lynch syndrome in patients with putative familial colorectal cancer. J Natl Compr Cancer Netw 9:1311–1320
Yurgelun MB, Hampel H (2018) Recent advances in Lynch syndrome: diagnosis, treatment, and cancer prevention. Am Soc Clin Oncol Educ Book 38:101–109
Wang T, Lee LH, Vyas M, Zhang L, Ganesh K, Firat C et al (2019) Colorectal carcinoma with double somatic mismatch repair gene inactivation: clinical and pathological characteristics and response to immune checkpoint blockade. Mod Pathol 32:1551–1562
Pearlman R, Haraldsdottir S, de la Chapelle A, Jonasson JG, Liyanarachchi S, Frankel WL et al (2019) Clinical characteristics of patients with colorectal cancer with double somatic mismatch repair mutations compared with Lynch syndrome. J Med Genet 56:462–470
Gupta S, Provenzale D, Llor X, Halverson AL, Grady W, Chung DC et al (2019) NCCN Guidelines Insights: Genetic/familial high-risk assessment: colorectal, version 2.2019. J Natl Compr Cancer Netw 17:1032–1041
Colella S, Shen L, Baggerly KA, Issa JP, Krahe R (2003) Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques 35:146–150
Deng G, Chen A, Hong J, Chae HS, Kim YS (1999) Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res 59:2029–2033
Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS (2002) Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer 86:574–579
Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A et al (2015) Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 17:251–264
Shen R, Seshan VE (2016) FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44:e131
Middha S, Zhang L, Nafa K, Jayakumaran G, Wong D, Kim HR et al (2017) Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol 1:1–17
Moran S, Arribas C, Esteller M (2016) Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 8:389–399
Shia J, Zhang L, Shike M, Guo M, Stadler Z, Xiong X et al (2013) Secondary mutation in a coding mononucleotide tract in MSH6 causes loss of immunoexpression of MSH6 in colorectal carcinomas with MLH1/PMS2 deficiency. Mod Pathol 26:131–138
Shia J, Stadler ZK, Weiser MR, Vakiani E, Mendelsohn R, Markowitz AJ et al (2015) Mismatch repair deficient-crypts in non-neoplastic colonic mucosa in Lynch syndrome: insights from an illustrative case. Fam Cancer 14:61–68
Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P et al (2013) Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45:136–144
Jansen AM, van Wezel T, van den Akker BE, Ventayol Garcia M, Ruano D, Tops CM et al (2016) Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet 24:1089–1092
Elsayed FA, Kets CM, Ruano D, van den Akker B, Mensenkamp AR, Schrumpf M et al (2015) Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet 23:1080–1084
Latournerie M, Jooste V, Cottet V, Lepage C, Faivre J, Bouvier AM (2008) Epidemiology and prognosis of synchronous colorectal cancers. Br J Surg 95:1528–1533
Lindberg LJ, Ladelund S, Bernstein I, Therkildsen C, Nilbert M (2019) Risk of synchronous and metachronous colorectal cancer: population-based estimates in Denmark with focus on non-hereditary cases diagnosed after age 50. Scand J Surg 108:152–158
Hu H, Chang DT, Nikiforova MN, Kuan SF, Pai RK (2013) Clinicopathologic features of synchronous colorectal carcinoma: a distinct subset arising from multiple sessile serrated adenomas and associated with high levels of microsatellite instability and favorable prognosis. Am J Surg Pathol 37:1660–1670
Roth RM, Haraldsdottir S, Hampel H, Arnold CA, Frankel WL (2016) Discordant mismatch repair protein immunoreactivity in Lynch syndrome-associated neoplasms: a recommendation for screening synchronous/metachronous neoplasms. Am J Clin Pathol 146:50–56
Nakano K, Yamamoto H, Fujiwara M, Koga Y, Tsuruta S, Ihara E et al (2018) Clinicopathologic and molecular characteristics of synchronous colorectal carcinoma with mismatch repair deficiency. Am J Surg Pathol 42:172–182
Dowty JG, Win AK, Buchanan DD, Lindor NM, Macrae FA, Clendenning M et al (2013) Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat 34:490–497
Moller P, Seppala TT, Bernstein I, Holinski-Feder E, Sala P, Gareth Evans D et al (2018) Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 67:1306–1316
Yurgelun MB, Allen B, Kaldate RR, Bowles KR, Judkins T, Kaushik P et al (2015) Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology 149:604–613 e20
Ring KL, Bruegl AS, Allen BA, Elkin EP, Singh N, Hartman AR et al (2016) Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Mod Pathol 29:1381–1389
Stoffel EM, Koeppe E, Everett J, Ulintz P, Kiel M, Osborne J et al (2018) Germline genetic features of young individuals with colorectal cancer. Gastroenterology 154:897–905 e1
Ten Broeke SW, van der Klift HM, Tops CMJ, Aretz S, Bernstein I, Buchanan DD et al (2018) Cancer risks for PMS2-associated Lynch syndrome. J Clin Oncol 36:2961–2968
Dominguez-Valentin M, Sampson JR, Seppala TT, Ten Broeke SW, Plazzer JP, Nakken S et al (2020) Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med 22:15–25
Win AK, Jenkins MA, Dowty JG, Antoniou AC, Lee A, Giles GG et al (2017) Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomark Prev 26:404–412
Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C et al (2019) Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364:485–491
Chen W, Pearlman R, Hampel H, Pritchard CC, Markow M, Arnold C et al (2020) MSH6 immunohistochemical heterogeneity in colorectal cancer: comparative sequencing from different tumor areas. Hum Pathol 96:104–111
Lee SC, Guo JY, Lim R, Soo R, Koay E, Salto-Tellez M et al (2005) Clinical and molecular characteristics of hereditary non-polyposis colorectal cancer families in Southeast Asia. Clin Genet 68:137–145
Wei W, Liu F, Liu L, Li Z, Zhang X, Jiang F et al (2011) Distinct mutations in MLH1 and MSH2 genes in hereditary non-polyposis colorectal cancer (HNPCC) families from China. BMB Rep 44:317–322
Yilmaz A, Mirili C, Bilici M, Tekin SB (2020) Colorectal cancer in Lynch syndrome associated with PMS2 and MSH6 mutations. Int J Colorectal Dis 35:351–353
Kloor M, Huth C, Voigt AY, Benner A, Schirmacher P, von Knebel DM et al (2012) Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: a pathological study. Lancet Oncol 13:598–606
Brand RE, Dudley B, Karloski E, Das R, Fuhrer K, Pai RK et al (2020) Detection of DNA mismatch repair deficient crypts in random colonoscopic biopsies identifies Lynch syndrome patients. Fam Cancer 19:169–175
Yurgelun MB, Goel A, Hornick JL, Sen A, Turgeon DK, Ruffin MT et al (2012) Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev Res (Phila) 5:574–582
Cancer Genome Atlas N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337
Buchanan DD, Stewart JR, Clendenning M, Rosty C, Mahmood K, Pope BJ et al (2018) Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med 20:890–895
Yamaguchi K, Shimizu E, Yamaguchi R, Imoto S, Komura M, Hatakeyama S et al (2019) Development of an MSI-positive colon tumor with aberrant DNA methylation in a PPAP patient. J Hum Genet 64:729–740
Pai RK, Bettington M, Srivastava A, Rosty C (2019) An update on the morphology and molecular pathology of serrated colorectal polyps and associated carcinomas. Mod Pathol 32:1390–1415
Hampel H, Pearlman R, Beightol M, Zhao W, Jones D, Frankel WL et al (2018) Assessment of tumor sequencing as a replacement for Lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 4:806–813
Acknowledgements
This study was supported in part by National Cancer Institute (US) Grant P30 C008748 and by the Romeo Milio Lynch Syndrome Foundation.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vyas, M., Firat, C., Hechtman, J.F. et al. Discordant DNA mismatch repair protein status between synchronous or metachronous gastrointestinal carcinomas: frequency, patterns, and molecular etiologies. Familial Cancer 20, 201–213 (2021). https://doi.org/10.1007/s10689-020-00210-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-020-00210-4