Abstract
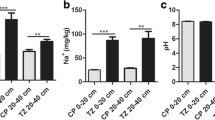
Salinity is a major constraint to the sustainability and expansion of maize cultivation. Plant salt tolerance is a quantitative trait controlled by multiple genes. In the present study, we constructed a high density genetic map based on high quality SNP markers from 161 F2:5 recombinant inbred line populations derived from the cross between two maize inbred lines contrasting in salinity tolerance. QTL analysis was conducted in saline field and the hydroponic culture. For saline field, field germination rate and field salt tolerance ranking were used as salinity tolerance indicators to conduct QTL detection. For hydroponic culture, salt tolerance ranking, shoot fresh weight, shoot dry weight, tissue water content, shoot Na+ concentration, shoot K+ concentration, and shoot K+/Na+ ratio were used. Through unconditional QTL analysis, we detected 20 additive and nine epistatic QTLs, of which 12 and two showed significant QTL by treatment (Q × T) interaction effects, respectively. Moreover, the use of conditional analysis model allowed us to detect nine conditional QTLs. The QTLs were mainly clustered on chromosomes 1, 3 and 5. The five unconditional and three conditional QTLs reported here could individually explain more than 20 % of the phenotypic variation. The QTLs identified here could be helpful to improve salt tolerance in maize by marker-assisted selection and shed new light on understanding the genetic basis of salt tolerance in maize.

Similar content being viewed by others
Abbreviations
- RIL:
-
Recombinant inbred line
- FGR:
-
Field germination rate
- FSTR:
-
Field salt tolerance ranking
- STR:
-
Salt tolerance ranking
- SFW:
-
Shoot fresh weight
- SDW:
-
Shoot dry weight
- TWC:
-
Tissue water content
- SNC:
-
Shoot Na+ concentration
- SKC:
-
Shoot K+ concentration
- SKN:
-
Shoot K+/Na+ ratio
- Q × T :
-
QTL by treatment interaction effects
- MAS:
-
Marker-assisted selection
- N:
-
Normal treatment
- S:
-
160 mM NaCl treatment
References
Cao G, Zhu J, He C, Gao Y, Yan J, Wu P (2001) Impact of epistasis and QTL × environment interaction on the developmental behavior of plant height in rice (Oryza sativa L.). Theor Appl Genet 103:153–160. doi:10.1007/s001220100536
Cui F, Li J, Ding AM, Zhao CH, Wang L, Wang XQ, Li SS, Bao YG, Li XF, Feng DS, Kong LR, Wang HG (2011) Conditional QTL mapping for plant height with respect to the length of the spike and internode in two mapping populations of wheat. Theor Appl Genet 122:1517–1536. doi:10.1007/s00122-011-1551-6
Cui F, Zhao CH, Li J, Ding AM, Li XF, Bao YG, Li JM, Ji J, Wang HG (2013) Kernel weight per spike: what contributes to it at the individual QTL level? Mol Breed 31:265–278. doi:10.1007/s11032-012-9786-8
DeRose-Wilson L, Gaut BS (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3:43–52. doi:10.1038/Nrg703
DeRose-Wilson L, Gaut BS (2011) Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS ONE 6:e22832. doi:10.1371/journal.pone.0022832
Fan CC, Yu XQ, Xing YZ, Xu CG, Luo LJ, Zhang QF (2005) The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor Appl Genet 110:1445–1452. doi:10.1007/s00122-005-1975-y
Ganal MW, Durstewitz G, Polley A, Berard A, Buckler ES, Charcosset A, Clarke JD, Graner EM, Hansen M, Joets J, Le Paslier MC, McMullen MD, Montalent P, Rose M, Schon CC, Sun Q, Walter H, Martin OC, Falque M (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6:e28334. doi:10.1371/journal.pone.0028334
Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet 121:877–894. doi:10.1007/s00122-010-1357-y
Ghomi K, Rabiei B, Sabouri H, Sabouri A (2013) Mapping QTLs for traits related to salinity tolerance at seedling stage of rice (Oryza sativa L.): an agrigenomics study of an Iranian rice population. OMICS 17:242–251. doi:10.1089/omi.2012.0097
Guo Y, Kong FM, Xu YF, Zhao Y, Liang X, Wang YY, An DG, Li SS (2012) QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor Appl Genet 124:851–865. doi:10.1007/s00122-011-1749-7
Jiang H, Jiang L, Guo LB, Gao ZY, Zeng DL, Zhu LH, Liang GH, Qian Q (2008) Conditional and unconditional mapping of quantitative trait loci underlying plant height and tiller number in rice (Oryza sativa L.) grown at two nitrogen levels. Prog Nat Sci 18:1539–1547. doi:10.1016/j.pnsc.2008.05.025
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenic 12:172–175
Lee GJ, Carter TE Jr, Villagarcia MR, Li Z, Zhou X, Gibbs MO, Boerma HR (2004) A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor Appl Genet 109:1610–1619. doi:10.1007/s00122-004-1783-9
Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, Lee KS, Eun MY (2006) Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol Cells 21:192–196
Li HH, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. doi:10.1534/genetics.106.066811
Li HH, Ribaut JM, Li ZL, Wang JK (2008a) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260. doi:10.1007/s00122-007-0663-5
Li YL, Dong YB, Cui DQ, Wang YZ, Liu YY, Wei MG, Li XH (2008b) The genetic relationship between popping expansion volume and two yield components in popcorn using unconditional and conditional QTL analysis. Euphytica 162:345–351. doi:10.1007/s10681-007-9513-4
Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108:253–260. doi:10.1007/s00122-003-1421-y
Liu G, Yang J, Xu H, Zhu J (2007) Influence of epistasis and QTL × environment interaction on heading date of rice (Oryza sativa L.). J Genet Genomics 34:608–615. doi:10.1016/S1673-8527(07)60069-1
Ma XQ, Tang JH, Teng WT, Yan JB, Meng YJ, Li JS (2007) Epistatic interaction is an important genetic basis of grain yield and its components in maize. Mol Breed 20:41–51. doi:10.1007/s11032-006-9071-9
Mano Y, akeda TK (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L). Euphytica 94:263–272. doi:10.1023/A:968207362
Marza F, Bai GH, Carver BF, Zhou WC (2006) Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor Appl Genet 112:688–698. doi:10.1007/s00122-005-0172-3
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. doi:10.1146/annurev.arplant.59.032607.092911
Prasad SR, Bagali PG, Hittalmani S, Shashidhar HE (2000) Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr Sci India 78:162–164
Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusic D, Waterman E, Weyen J, Schondelmaier J, Habash DZ, Farmer P, Saker L, Clarkson DT, Abugalieva A, Yessimbekova M, Turuspekov Y, Abugalieva S, Tuberosa R, Sanguineti MC, Hollington PA, Aragues R, Royo A, Dodig D (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110:865–880. doi:10.1007/s00122-004-1902-7
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146. doi:10.1038/ng1643
Rodriguez-Navarro A, Rubio F (2006) High-affinity potassium and sodium transport systems in plants. J Exp Bot 57:1149–1160. doi:10.1093/Jxb/Erj068
Saghaimaroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley—mendelian inheritance, chromosomal location, and population-dynamics. Proc Natl Acad Sci USA 81:8014–8018. doi:10.1073/pnas.81.24.8014
Serrano R (1996) Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol 165:1–52. doi:10.1016/S0074-7696(08)62219-6
Shen XL, Zhang TZ, Guo WZ, Zhu XF, Zhang XY (2006) Mapping fiber and yield QTLs with main, epistatic, and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Sci 46:61–66. doi:10.2135/scopsci2005.0056
Sun XY, Wu K, Zhao Y, Kong FM, Han GZ, Jiang HM, Huang XJ, Li RJ, Wang HG, Li SS (2009) QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 165:615–624. doi:10.1007/s10681-008-9794-2
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527. doi:10.1093/Aob/Mcg058
Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM (2010) Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice 3:148–160. doi:10.1007/s12284-010-9053-8
Wang Z, Wu X, Ren Q, Chang X, Li R, Jing R (2010) QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum L.). Euphytica 174:447–458. doi:10.1007/s10681-010-0166-3
Wang Z, Cheng J, Chen Z, Huang J, Bao Y, Wang J, Zhang H (2012) Identification of QTLs with main, epistatic and QTL × environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor Appl Genet 125:807–815. doi:10.1007/s00122-012-1873-z
Wu JX, Jenkins JN, McCarty JC, Zhu J (2004) Genetic association of yield with its component traits in a recombinant inbred line population of cotton. Euphytica 140:171–179. doi:10.1007/s10681-004-2897-5
Xu YF, An DG, Liu DC, Zhang AM, Xu HX, Li B (2012) Mapping QTLs with epistatic effects and QTL × treatment interactions for salt tolerance at seedling stage of wheat. Euphytica 186:233–245. doi:10.1007/s10681-012-0647-7
Xu Y, Li S, Li L, Zhang X, Xu H, An D, Igartua E (2013) Mapping QTLs for salt tolerance with additive, epistatic and QTL × treatment interaction effects at seedling stage in wheat. Plant Breed 132:276–283. doi:10.1111/pbr.12048
Xu Y, Wang R, Tong Y, Zhao H, Xie Q, Liu D, Zhang A, Li B, Xu H, An D (2014) Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Theor Appl Genet 127:59–72. doi:10.1007/s00122-013-2201-y
Yan JQ, Zhu J, He CX, Benmoussa M, Wu P (1998) Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor Appl Genet 97:267–274. doi:10.1007/s001220050895
Yan JB, Tang H, Huang YQ, Shi YG, Li JS, Zheng YL (2003) Dynamic analysis of QTL for plant height at different developmental stages in maize (Zea mays L.). Chin Sci Bull 48:2601–2607. doi:10.1360/03wc0044
Yang J, Hu CC, Hu H, Yu RD, Xia Z, Ye XZ, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723. doi:10.1093/bioinformatics/btm494
Zhang KP, Tian JC, Zhao L, Wang SS (2008) Mapping QTLs with epistatic effects and QTL × environment interactions for plant height using a doubled haploid population in cultivated wheat. J Genet Genomics 35:119–127. doi:10.1016/S1673-8527(08)60017-X
Zhang H, Cui F, Wang L, Li J, Ding AM, Zhao CH, Bao YG, Yang QP, Wang HG (2013) Conditional and unconditional QTL mapping of drought-tolerance-related traits of wheat seedling using two related RIL populations. J Genet 92:213–231. doi:10.1007/s12041-013-0253-z
Zhao JY, Becker HC, Zhang DQ, Zhang YF, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113:33–38. doi:10.1007/s00122-006-0267-5
Zhu J (1995) Analysis of conditional genetic-effects and variance-components in developmental genetics. Genetics 141:1633–1639
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71. doi:10.1016/S1360-1385(00)01838-0
Acknowledgments
The authors thank the China National Science Foundation (Grant No. 31201214) and the National Sci-Tech Support program (Grant No.2013BAD05B01) for providing funds for carrying out this research work. We are very grateful to Dr. Jiping Zhao, Manager-Developmental Biology, Ball Horticultural Company, United States, for thorough English editing of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Dezhou Cui and Dandan Wu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cui, D., Wu, D., Somarathna, Y. et al. QTL mapping for salt tolerance based on snp markers at the seedling stage in maize (Zea mays L.). Euphytica 203, 273–283 (2015). https://doi.org/10.1007/s10681-014-1250-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1250-x