Abstract
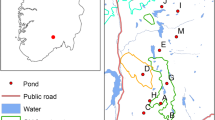
The reticulated flatwoods salamander (Ambystoma bishopi), an endangered species endemic to the longleaf-pine savanna ecosystem of the southeastern U.S., persists in a small number of remnant habitat patches. Breeding ponds and associated populations are threatened by habitat loss, degradation, and fragmentation stemming from fire suppression and land conversion. Understanding influences on population dynamics and genetic diversity will help inform recovery efforts for this and other pond-breeding amphibians. We used 9 microsatellite loci to characterize population structure, migration, and genetic diversity of juvenile (larvae and metamorphs) A. bishopi (n = 607) sampled from thirteen breeding ponds across two breeding seasons. Temporal genetic variation between annual cohorts was minimal compared to spatial variation among locations. The primary genetic subdivision was between two regional groups of breeding ponds (i.e. metapopulations), which were located more than 10 km from each other. Yet even within metapopulations, genetic isolation-by-distance was pronounced, and estimated migration rates among ponds separated by > 400 m were very low. Genetic diversity was positively correlated with the area of suitable breeding habitat in a pond and negatively correlated with distance to other occupied ponds. The effective number of breeders typically was < 30 individuals per year per pond, and all populations showed signs of having been through a severe demographic bottleneck. Small and variable local population sizes suggest the importance of metapopulation dynamics among ponds for maintaining regional persistence and genetic diversity. Such dynamics may be easily disrupted for A. bishopi, so maintaining and restoring connectivity appears crucial for long-term conservation.





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available on Dryad. Data regarding microsatellite primer sequences can be found in supplementary information. Any other data, excluding exact locations of ponds, which cannot be publicly shared due to the species’ protected status, can be obtained from the corresponding author.
Code availability
Custom code can be found in Supplementary Materials.
Change history
15 May 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10592-021-01367-w
References
Baber MJ, Fleishman E, Babbitt KJ, Tarr TL (2004) The relationship between wetland hydroperiod and nestedness patterns in assemblages of larval amphibians and predatory macroinvertebrates. Oikos 107(1):16–27
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11:155–165
Bishop DC, Palis JG, Enge KM, Printiss DJ, Stevenson DJ (2006) Capture rate, body size, and survey recommendations for larval Ambystoma cingulatum (flatwoods salamanders). Southeast Nat 5:9–16
Brooks GC (2020) On the use of demographic models to inform amphibian conservation and management: a case study of the Reticulated Flatwoods Salamander. Dissertation, Virginia Tech
Brooks GC, Smith JA, Frimpong EA, Gorman TA, Chandler HC, Haas CA (2019a) Indirect connectivity estimates of amphibian breeding wetlands from spatially explicit occupancy models. Aquat Conserv 29(11):1815–1825
Brooks GC, Smith JA, Gorman TA, Haas CA (2019b) Discerning the environmental drivers of annual migrations in an endangered amphibian. Copeia 107:270–276. https://doi.org/10.1643/CH-18-068
Brooks GC, Gorman TA, Jiao Y, Haas CA (2020) Reconciling larval and adult sampling methods to model growth across life-stages. PLoS ONE 15(18):e0237737. https://doi.org/10.1371/journal.pone.0237737
Caye K, Jay F, Michel O, Francois O (2016) TESS3: fast inference of spatial population structure and genome scans for selection. Mol Ecol Resour 16:540–548
Chandler HC, Haas CA, Gorman TA (2015) The effects of habitat structure on winter aquatic invertebrate and amphibian communities in pine flatwoods wetlands. Wetlands 35:1201–1211
Chandler HC, Rypel AL, Jiao Y, Haas CA, Gorman TA (2016) Hindcasting historical breeding conditions of an endangered salamander in ephemeral wetlands of the Southeastern USA: implications of climate change. PLoS ONE 11(2):e0150169
Chandler HC, McLaughlin DL, Gorman TA, McGuire KJ, Feaga JB, Haas CA (2017) Drying rates of ephemeral wetlands: implications for breeding amphibians. Wetlands 37:545–557
Clipp H, Anderson J (2014) Environmental and anthropogenic factors influencing salamanders in riparian forests: a review. Forests 5:2679–2702
Cushman S (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Cons 128:231–240
Douglas ME, Monroe BL (1981) A comparative study of topographical orientation in Ambystoma (Amphibia: Caudata). Copeia 1981:460–463
Eigenbrod F, Hecnar S, Fahrig L (2008) The relative effects of road traffic and forest cover on anuran populations. Biol Conserv 141:35–46
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Evol Syst 24:217–242
Erwin KJ, Chandler HC, Palis JG, Gorman TA, Haas CA (2016) Herpetofaunal communities in ephemeral wetlands embedded within longleaf pine flatwoods of the Gulf Coastal Plain. Southeast Nat 15(3):431–447
Excoffier L, Lischer HEL (2010) ARLEQUIN suite, version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fahrig L, Merriam G (1994) Conservation of fragmented populations. Conserv Biol 8:50–59
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16(6):1149–1166
Fischer M, Matthies D (1998) Effects of population size on performance in the rare plant Gentianella germanica. J Ecol 86:195–204
Franklin IR, Frankham R (1998) How large must populations be to retain evolutionary potential? Anim Conserv 1:69–73
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Gamble LR (2004) Landscape-level population structure and local variability in marbled salamanders (Ambystoma opacum) of western Massachusetts: applied lessons from metapopulation theory. Thesis, University of Massachusetts-Amherst
Gamble L, McGarifal K, Compton B (2007) Fidelity and dispersal in the pond-breeding amphibian, Ambystoma opacum: implication for spatio-temporal population dynamics and conservation. Biol Conserv 139:247–257
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Gibbs JP (1998) Genetic structure of redback salamander Plethodon cinereus populations in continuous and fragmented forests. Biol Conserv 86:77–81
Gibbs J (2000) Wetland loss and biodiversity conservation. Conserv Biol 14:314–317
Gillespie G (2001) The role of introduced trout in the decline of the spotted tree frog (Litoria spenceri) in South-eastern Australia. Biol Conserv 100:87–198
Gonzalez A, Lawton JH, Gilbert FS, Blackburn TM, Evans-Freke I (1998) Metapopulation dynamics, abundance, and distribution in a microecosystem. Science 281:2045–2047
Gorman TA, Haas CA, Bishop DC (2009) Factors related to occupancy of breeding wetlands by flatwoods salamander larvae. Wetlands 29(1):323–329
Gorman TA, Haas CA, Himes JG (2013) Evaluating methods to restore amphibian habitat in fire-suppressed pine flatwoods wetlands. Fire Ecol 9:96–109
Gorman TA, Powell SD, Jones KC, Haas CA (2014) Microhabitat characteristics of egg deposition sites used by reticulated flatwoods salamanders. Herpetol Conserv Biol 9:543–550
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Goudet J (2002) FSTAT: A program to estimate and test gene diversities and fixation indices, Version 2.9.3.2
Graeter GJ, Rothermel BB, Gibbons JW (2008) Habitat selection and movement of pond-breeding amphibians in experimentally fragmented pine forests. J Wildl Manag 72(2):473–482
Greenwald KR (2010) Genetic data in population viability analysis: case studies with ambystomatid salamanders. Anim Conserv 13:115–122
Guerry AD, Hunter ML Jr (2002) Amphibian distributions in a landscape of forests and agriculture: an examination of landscape composition and configuration. Conserv Biol 16:745–754
Gulve PS (1994) Distribution and extinction patterns within a northern metapopulation of the pool frog, Rana lessonae. Ecology 75:1357–1367
Hale ML, Burg TM, Steeves TE (2012) Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 7(9):e45170
Harper E, Semlitsch R (2007) Density dependence in the terrestrial life history stage of two anurans. Oecologia 153:879–889
Jacquemyn H, Brys R, Hermy M (2002) Patch occupancy, population size and reproductive success of a forest herb (Primula elatior) in a fragmented landscape. Oecologia 130:617–625
Jehle R, Wilson GA, Arntzen JW, Burke T (2005) Contemporary gene flow and the spatio-temporal genetic structure of subdivided newt populations (Triturus cristatus, T. marmoratus). J Evol Biol 18:619–628
Johansson M, Primmer CR, Merilä J (2007) Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog (Rana temporaria). Mol Ecol 16(13):2693–2700
Jones O, Wang J (2010) Molecular marker-based pedigrees for animal conservation biologists. Anim Conserv 13:26–34
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106(4):625–632
Kershenbaum A, Blank L, Sinai I, Merilä J, Blaustein L, Templeton AR (2014) Landscape influences on dispersal behaviour: a theoretical model and empirical test using the fire salamander, Salamandra infraimmaculata. Oecologia 175:509–520
Kinkead KE, Abbott AG, Otis DL (2007) Genetic variation among Ambystoma breeding populations on the Savannah river site. Conserv Genet 8:281–292
Kleeberger SR, Werner JR (1983) Post-breeding migration and summer movement of Ambystoma maculatum. J Herpetol 17:176–177
Mitchell J, Gibbons W (2010) Salamanders of the Southeast. University of Georgia Press, Athens
Nei M, Chakravarti A (1977) Drift variances of Fst and Gst statistics obtained from a finite number of isolated populations. Theor Popul Biol 11:307–325
O’Connell KA, Mulder KP, Maldonado J, Currie KL, Ferraro DM (2019) Sampling related individuals within ponds biases estimates of population structure in a pond-breeding amphibian. Ecol Evolut 9(6):3620–3636
Palis J (1995) Larval growth, development, and metamorphosis of Ambystoma cingulatum on the Gulf Coastal Plain of Florida. Florida Scientist 58:352–358
Palis J (1997a) Breeding migration of Ambystoma cingulatum in Florida. J Herpetol 31(1):71–78
Palis J (1997b) Distribution, habitat, and status of the Flatwoods Salamander (Ambystoma cingulatum) in Florida, USA. Herpetol Natural History 5:53–65
Palis JG, Aresco MJ, Kilpatrick S (2006) Breeding biology of a Florida population of Ambystoma cingulatum (Flatwoods salamander) during a drought. Southeast Nat 5(1):1–8
Pauly G, Piskurek O, Shaffer H (2007) Phylogeographic concordance in the southeastern United States: the flatwoods salamander, Ambystoma cingulatum, as a test case. Mol Ecol 16:415–429
Pearman PB, Garner TWJ (2006) Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecol Lett 8:401–408
Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carrillo C, Pauli JN, Palsbøll PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21(14):3403–3418
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in terrestrial woodland salamander. Mol Ecol 23:2402–2413
Peterman WE, Anderson TL, Ousterhout BH, Drake DL, Semlitsch RD, Eggert LS (2015) Differential dispersal shapes population structure and patterns of genetic differentiation in two sympatric pond breeding salamanders. Conserv Genet 16:59–69
Peterman W, Brocato ER, Semlitsch RD, Eggert LS (2016) Reducing bias in population and landscape genetic inferences: the effects of sampling related individuals and multiple life stages. PeerJ 4:e1813
Polo-Cavia N, Gomez-Mestre I (2014) Learned recognition of introduced predators determines survival of tadpole prey. Funct Ecol 28:432–439
Powell SD, Gorman TA, Haas CA (2015) Nightly movements and use of burrows by Reticulated Flatwoods Salamanders (Ambystoma bishopi) in breeding wetlands. Florida Sci 78(3/4):149–155
Puechmaille SJ (2016) The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627
Purrenhage JL, Niewiarowski H, Moore FB-G (2009) Population structure of spotted salamanders (Ambystoma maculatum) in fragmented landscapes. Mol Ecol 18:235–247
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Reynolds J, Weir BS, Cockerham CC (1983) Estimation for the coancestry coefficient: basis for the short-term genetic distance. Genetics 105:767–779
Richardson JL (2012) Divergent landscape effects on population connectivity in two co-occurring amphibian species. Mol Ecol 21:4437–4451
Richardson JL, Urban MC (2013) Strong selection barriers explain microgeographic adaptation in wild salamander populations. Evolution 67(6):1729–1740
Riley S, Busteed GT, Kats LB, Vandergon TL, Lee LFS, Dagit RG, Kerby JL, Fisher RN, Sauvajot RM (2005) Effects of urbanization on the distribution and abundance of amphibians and invasive species in Southern California streams. Conserv Biol 19(6):1894–1907
Rothermel BB, Semlitsch RD (2002) An experimental investigation of landscape resistance of forest versus old-field habitats to emigrating juvenile amphibians. Conserv Biol 16:619–629
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145(4):1219–1228
Scott DE, Komoroski MJ, Croshaw DA, Dixon PM (2013) Terrestrial distribution of pond-breeding salamanders around an isolated wetland. Ecology 94:2537–2546
Semlitsch RD (1998) Biological delineation of terrestrial buffer zones for pond-breeding salamanders. Conserv Biol 5:1113–1119
Semlitsch RD, Walls SC, Barichivich WJ, O’Donnell KM (2017) Extinction debt as a driver of amphibian declines: an example with imperiled flatwoods salamanders. J Herpetol 51(1):12–18
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetic Resour 68:259–260
Spear SF, Peterson CS, Matocq MD, Storfer A (2005) Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum). Mol Ecol 14:2553–2564
Spear SF, Peterson CR, Matocq MD, Storger A (2006) Molecular evidence for the historical and recent population size reduction of tiger salamanders (Ambystoma tigrinum) in Yellowstone National Park. Conserv Genet 7(4):605–611
Storfer A, Mech SG, Reudink MW, Lew K (2014) Inbreeding and strong population subdivision in an endangered salamander. Conserv Genet 15:137–151
Stuart S, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW (2004) Status and trends of amphibian declines and extinctions worldwide. Science 306:1783–1786
Szczecińska M, Sramko G, Wolosz K, Sawicki J (2016) Genetic diversity and population structure of the rare and endangered plant species Pulsatilla patens (L.) Mill in east central Europe. PLoS ONE 11(3):e0151730
Trenham PC, Koenig WD, Shaffer HB (2001) Spatially autocorrelated demography and interpond dispersal in the salamander Ambystom californiense. Ecology 82(12):3519–3530
United States Department of the Interior, Fish and Wildlife Service (2009) Endangered and threatened wildlife and plants; determination of endangered status for the reticulated flatwoods salamander; designation of the critical habitat for frosted flatwoods salamander and reticulated flatwoods salamander; final rule. Fed Reg 64:6699–6774
Van Lear DH, Carroll WD, Kapeluck PR, Johnson R (2005) History and restoration of the longleaf pine-grassland ecosystem: implications for species at risk. For Ecol Manage 211(1–2):150–165
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538
Walls SC, Barichivich WJ, Brown ME (2013) Drought, deluge and declines: the impact of precipitation extremes on amphibians in a changing climate. Biology 2:399–418
Wang J (2004) Sibship reconstruction from genetic data with typing errors. Genetics 166:1963–1979
Wang IJ, Shaffer HB (2017) Population genetic and field-ecological analyses return similar estimates of dispersal over space and time in an endangered amphibian. Evol Appl 10(6):630–639
Wang IJ, Johnson JR, Johnson BB, Shaffer HB (2011) Effective population size is strongly correlated with breeding pond size in the endangered California tiger salamander, Ambysotma californiense. Conserv Genet 12(4):911–920
Waples RS, Anderson EC (2017) Purging putative siblings from population genetic data sets: a cautionary review. Mol Ecol 26:1211–1224
Waples RS, Do C (2008) LDNe: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8(4):753–756
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3(3):244–262
Waples RS, Luikart G, Faulkner JR, Tallmon DA (2013) Simple life-history traits explain key effective population size ratios across diverse taxa. Proc R Soc B. https://doi.org/10.1098/rspb.2013.1339
Wendt A (2017) A population genetic investigation of the reticulated flatwoods salamander (Ambystoma bishopi) on Eglin Air Force Base. Thesis, Georgia Southern University
Whiteley AR, McGarigal K, Schwartz MK (2014) Pronounced differences in genetic structure despite overall similarity for two Ambystoma salamanders in the same landscape. Conserv Genet 15:573–591
Williams ST (2019) Immune Gene Diversity and Population Structure of Reticulated Flatwoods Salamander (Ambystoma bishopi). Master’s Thesis, Louisiana State University, Baton Rouge, Louisiana
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163(3):1177–1191
Whitlock MC, Barton NH (1997) The effective size of a subdivided population. Genetics 146(1):427–441
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11(10):413–418
Zamudio KR, Wieczorek AM (2007) Fine-scale spatial genetic structure and dispersal among spotted salamander (Ambystoma maculatum) breeding populations. Mol Ecol 16:257–274
Acknowledgements
We express our sincerest thanks to G. Brooks, K. Jones, B. Rincon, V. Porter, H. Chandler, S. Goodman, A. Hillman, J. Newman, K. Erwin, J. Newton, S. Konkolics, C. Abeles, (A) Perez-Umphrey, T. Williams, J. Da Silva Neto, J. Hefner, T. Mammone, (B) Moore, (A) Saenger, and other members of the Eglin field crew for their hard work in providing samples and metadata for this analysis. G. Brooks, N. Caruso, J. Eschenroeder, S. Harrison, (B) Scott, J. Smith and G. Strickland assisted in various invaluable ways with lab work and data analysis. Manuscript drafts have benefitted from the helpful comments of (C) Abeles, S. Harrison, L. McBrayer, and S. Taylor. Funding and support was provided by the Natural Resources Branch of Eglin Air Force Base, Department of Defense Legacy Resource Management Program, Hurlburt Field (USAF), U.S. Fish and Wildlife Service, Aquatic Habitat Restoration/Enhancement Program of the Florida Fish and Wildlife Conservation Commission, the Department of Fish and Wildlife Conservation at Virginia Tech, the USDA National Institute of Food and Agriculture, McIntire Stennis project 1006328, and the Graduate Student Organization of Georgia Southern University. We sincerely appreciate the commitment and support from dedicated colleagues at the agencies mentioned above who have gone above and beyond to support our work and the conservation of this species.
Funding
Funding and support was provided by Natural Resources Branch of Eglin Air Force Base, Department of Defense Legacy Resource Management Program, Hurlburt Field (USAF), U.S. Fish and Wildlife Service, Aquatic Habitat Restoration/Enhancement (AHRE) Program of Florida Fish and Wildlife Conservation Commission, Department of Fish and Wildlife Conservation at Virginia Tech, and GSU Graduate Student Organization.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest within this publication.
Consent for publication
All authors give consent for publication.
Consent to participate
Not applicable.
Ethical approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wendt, A., Haas, C.A., Gorman, T. et al. Metapopulation genetics of endangered reticulated flatwoods salamanders (Ambystoma bishopi) in a dynamic and fragmented landscape. Conserv Genet 22, 551–567 (2021). https://doi.org/10.1007/s10592-021-01360-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01360-3