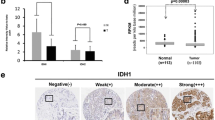
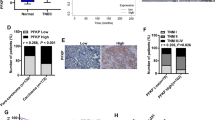
Abstract
Background
Breast cancer (BC) is regarded as one of the most common cancers diagnosed among the female population and has an extremely high mortality rate. It is known that Fibronectin 1 (FN1) drives the occurrence and development of a variety of cancers through metabolic reprogramming. Aspartic acid is considered to be an important substrate for nucleotide synthesis. However, the regulatory mechanism between FN1 and aspartate metabolism is currently unclear.
Methods
We used RNA sequencing (RNA seq) and liquid chromatography-mass spectrometry to analyze the tumor tissues and paracancerous tissues of patients. MCF7 and MDA-MB-231 cells were used to explore the effects of FN1-regulated aspartic acid metabolism on cell survival, invasion, migration and tumor growth. We used PCR, Western blot, immunocytochemistry and immunofluorescence techniques to study it.
Results
We found that FN1 was highly expressed in tumor tissues, especially in Lumina A and TNBC subtypes, and was associated with poor prognosis. In vivo and in vitro experiments showed that silencing FN1 inhibits the activation of the YAP1/Hippo pathway by enhancing YAP1 phosphorylation, down-regulates SLC1A3-mediated aspartate uptake and utilization by tumor cells, inhibits BC cell proliferation, invasion and migration, and promotes apoptosis. In addition, inhibition of FN1 combined with the YAP1 inhibitor or SLC1A3 inhibitor can effectively inhibit tumor growth, of which inhibition of FN1 combined with the YAP1 inhibitor is more effective.
Conclusion
Targeting the “FN1/YAP1/SLC1A3/Aspartate metabolism” regulatory axis provides a new target for BC diagnosis and treatment. This study also revealed that intratumoral metabolic heterogeneity plays an important role in the progression of different subtypes of breast cancer.






Similar content being viewed by others
Data availability
The RNA-seq data have been deposited in the NCBI GEO under accession number PRJNA962803, and the metabolomics data in this study can be obtained from the corresponding author on request.
Change history
28 December 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10549-023-07194-6
Abbreviations
- Asp:
-
Aspartate
- BC:
-
Breast cancer
- CCK-8:
-
Cell Counting Kit-8
- Co-IP:
-
Co-immunoprecipitation
- CRC:
-
Colorectal cancer
- EAAT1:
-
Excitatory amino acid transporter 1
- ECM:
-
Extracellular matrix
- FBS:
-
Fetal bovine serum
- FDR:
-
False discovery rate
- FN:
-
Fibronectin
- GLAST:
-
Glutamate/aspartate transporter
- GO:
-
Gene ontology
- HCA:
-
Hierarchical cluster analysis
- IF:
-
Immunofluorescence staining
- IHC:
-
Immunohistochemistry
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LC–MS:
-
Liquid chromatography–mass spectrometry
- NCCR:
-
National Central Cancer Registry
- OSR:
-
Overall survival rate
- qRT-PCR:
-
Reverse Transcriptase qPCR
- Q-TOF MS:
-
Quadrupole time-of-flight mass spectrometer
- RNA seq:
-
RNA sequencing
- SLC:
-
Solute carrier family
- SLC1A3:
-
Solute carrier family 1 member 3
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Lei S, Zheng R, Zhang S, Chen R, Wang S, Sun K et al (2021) Breast cancer incidence and mortality in women in China: temporal trends and projections to 2030. Cancer Biol Med. https://doi.org/10.20892/j.issn.2095-3941.2020.0523
Ren X (2021) Cancer immunology and immunotherapy. Cancer Biol Med 18:931–933
Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 321:288–300. https://doi.org/10.1001/jama.2018.19323
de la Cruz-Merino L, Palazón-Carrión N, Henao-Carrasco F, Nogales-Fernández E, Álamo-de la Gala M, Vallejo-Benítez A et al (2019) New horizons in breast cancer: the promise of immunotherapy. Clin Transl Oncol 21:117–125. https://doi.org/10.1007/s12094-018-1907-3
Hida K, Ishii G (2016) Editorial: targeting tumor microenvironment heterogeneity. Adv Drug Deliv Rev 99:139. https://doi.org/10.1016/j.addr.2016.03.004
Roulot A, Héquet D, Guinebretière JM, Vincent-Salomon A, Lerebours F, Dubot C, Rouzier R (2016) Tumoral heterogeneity of breast cancer. Ann Biol Clin (Paris) 74:653–660
Faubert B, Solmonson A, DeBerardinis RJ (2020) Metabolic reprogramming and cancer progression. Science 368:6487. https://doi.org/10.1126/science.aaw5473
Dey P, Kimmelman AC, DePinho RA (2021) Metabolic codependencies in the tumor microenvironment. Cancer Discov 11:1067–1081. https://doi.org/10.1158/2159-8290.CD-20-1211
Liu C, Li M, Dong ZX, Jiang D, Li X, Lin S et al (2021) Heterogeneous microenvironmental stiffness regulates pro-metastatic functions of breast cancer cells. Acta Biomater 131:326–340. https://doi.org/10.1016/j.actbio.2021.07.009
Evans KW, Yuca E, Scott SS, Zhao M, Paez Arango N, Cruz Pico CX et al (2021) Oxidative phosphorylation is a metabolic vulnerability in chemotherapy-resistant triple-negative breast cancer. Cancer Res 81:5572–5581. https://doi.org/10.1158/0008-5472.CAN-20-3242
Rossi M, Altea-Manzano P, Demicco M, Doglioni G, Bornes L, Fukano M et al (2022) PHGDH heterogeneity potentiates cancer cell dissemination and metastasis. Nature 605:747–753
Mao Y, Schwarzbauer JE (2005) Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol 24:389–399. https://doi.org/10.1016/j.matbio.2005.06.008
Speziale P, Arciola CR, Pietrocola G (2019) Fibronectin and its role in human infective diseases. Cells 8:1516. https://doi.org/10.3390/cells8121516
Cai X, Liu C, Zhang TN, Zhu YW, Dong X, Xue P (2018) Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J Cell Biochem 119:4717–4728. https://doi.org/10.1002/jcb.26651
Erdogan B, Ao M, White LM, Means AL, Brewer BM, Yang L et al (2017) Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J Cell Biol 216:3799–3816. https://doi.org/10.1083/jcb.201704053
Chen S, Chai X, Wu X (2022) Bioinformatical analysis of the key differentially expressed genes and associations with immune cell infiltration in development of endometriosis. BMC Genom Data 23:20. https://doi.org/10.1186/s12863-022-01036-y
Sun Y, Zhao C, Ye Y, Wang Z, He Y, Li Y et al (2020) High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol Lett 19:93–102. https://doi.org/10.3892/ol.2019.11088
Zhang Z, Peng Y, Dang J, Liu X, Zhu D, Zhang Y et al (2022) Identification of key biomarkers related to epithelial-mesenchymal transition and immune infiltration in ameloblastoma using integrated bioinformatics analysis. Oral Dis. https://doi.org/10.1111/odi.14173
Wang J, Li R, Li M, Wang C (2021) Fibronectin and colorectal cancer: signaling pathways and clinical implications. J Recept Signal Transduct Res 41:313–320. https://doi.org/10.1080/10799893.2020.1817074
Bao H, Huo Q, Yuan Q, Xu C (2021) Fibronectin 1: a potential biomarker for ovarian cancer. Dis Markers 2021:5561651. https://doi.org/10.1155/2021/5561651
Chen R, Chen L (2022) Solute carrier transporters: emerging central players in tumour immunotherapy. Trends Cell Biol 32:186–201. https://doi.org/10.1016/j.tcb.2021.08.002
Xu L, Chen J, Jia L, Chen X, Awaleh Moumin F, Cai J (2020) SLC1A3 promotes gastric cancer progression via the PI3K/AKT signalling pathway. J Cell Mol Med 24:14392–14404. https://doi.org/10.1111/jcmm.16060
Freidman N, Chen I, Wu Q, Briot C, Holst J, Font J et al (2020) Amino acid transporters and exchangers from the SLC1A family: structure, mechanism and roles in physiology and cancer. Neurochem Res 45:1268–1286. https://doi.org/10.1007/s11064-019-02934-x
Garcia-Bermudez J, Baudrier L, La K, Zhu XG, Fidelin J, Sviderskiy VO et al (2018) Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol 20:775–781. https://doi.org/10.1038/s41556-018-0118-z
Krall AS, Mullen PJ, Surjono F, Momcilovic M, Schmid EW, Halbrook CJ et al (2021) Asparagine couples mitochondrial respiration to ATF4 activity and tumor growth. Cell Metab 33:1013–26.e6. https://doi.org/10.1016/j.cmet.2021.02.001
Tajan M, Hock AK, Blagih J, Robertson NA, Labuschagne CF, Kruiswijk F et al (2018) A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Cell Metab 28:721–36.e6. https://doi.org/10.1016/j.cmet.2018.07.005
Di Benedetto G, Parisi S, Russo T, Passaro F (2021) YAP and TAZ mediators at the crossroad between metabolic and cellular reprogramming. Metabolites 11:154. https://doi.org/10.3390/metabo11030154
Raj N, Bam R (2019) Reciprocal crosstalk between YAP1/hippo pathway and the p53 family proteins: mechanisms and outcomes in cancer. Front Cell Dev Biol 7:159. https://doi.org/10.3389/fcell.2019.00159
Yamaguchi H, Taouk GM (2020) A potential role of YAP/TAZ in the interplay between metastasis and metabolic alterations. Front Oncol 10:928. https://doi.org/10.3389/fonc.2020.00928
Mohajan S, Jaiswal PK, Vatanmakarian M, Yousefi H, Sankaralingam S, Alahari SK et al (2021) Hippo pathway: regulation, deregulation and potential therapeutic targets in cancer. Cancer Lett 507:112–123. https://doi.org/10.1016/j.canlet.2021.03.006
Sun S, Irvine KD (2016) Cellular organization and cytoskeletal regulation of the hippo signaling network. Trends Cell Biol 26:694–704. https://doi.org/10.1016/j.tcb.2016.05.003
Yang CS, Stampouloglou E, Kingston NM, Zhang L, Monti S, Varelas X (2018) Glutamine-utilizing transaminases are a metabolic vulnerability of TAZ/YAP-activated cancer cells. EMBO Rep. https://doi.org/10.15252/embr.201643577
Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S et al (2015) A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun 6:10186. https://doi.org/10.1038/ncomms10186
Ye Y, Zhang R, Feng H (2020) Fibronectin promotes tumor cells growth and drugs resistance through a CDC42-YAP-dependent signaling pathway in colorectal cancer. Cell Biol Int 44:1840–1849. https://doi.org/10.1002/cbin.11390
Chong EY, Huang Y, Wu H, Ghasemzadeh N, Uppal K, Quyyumi AA et al (2015) Local false discovery rate estimation using feature reliability in LC/MS metabolomics data. Sci Rep 5:17221. https://doi.org/10.1038/srep17221
Yu L, Fernandez S, Brock G (2020) Power analysis for RNA-Seq differential expression studies using generalized linear mixed effects models. BMC Bioinform 21(1):198. https://doi.org/10.1186/s12859-020-3541-7
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG (2015) Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162:552–563
Bertero T, Oldham WM, Grasset EM, Bourget I, Boulter E, Pisano S et al (2019) Tumor-Stroma mechanics coordinate amino acid availability to sustain tumor growth and malignancy. Cell Metab 29:124–40.e10. https://doi.org/10.1016/j.cmet.2018.09.012
Jensen EC (2013) Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec (Hoboken) 296:378–381
Varghese F, Bukhari AB, Malhotra R, De A (2014) IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 9:e96801
Wei H, Wang F, Wang Y, Li T, Xiu P, Zhong J et al (2017) Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci 108:478–487
Abrahamsen B, Schneider N, Erichsen MN, Huynh TH, Fahlke C, Bunch L et al (2013) Allosteric modulation of an excitatory amino acid transporter: the subtype-selective inhibitor UCPH-101 exerts sustained inhibition of EAAT1 through an intramonomeric site in the trimerization domain. J Neurosci 33:1068–1087
Lin Y, Yang Z, Li J, Sun Y, Zhang X, Qu Z et al (2022) Effects of glutamate and aspartate on prostate cancer and breast cancer: a Mendelian randomization study. BMC Genomics 23:213. https://doi.org/10.1186/s12864-022-08442-7
Wu S, Zheng Q, Xing X, Dong Y, Wang Y, You Y et al (2018) Matrix stiffness-upregulated LOXL2 promotes fibronectin production, MMP9 and CXCL12 expression and BMDCs recruitment to assist pre-metastatic niche formation. J Exp Clin Cancer Res 37:99. https://doi.org/10.1186/s13046-018-0761-z
Moroz A, Delella FK, Lacorte LM, Deffune E, Felisbino SL (2013) Fibronectin induces MMP2 expression in human prostate cancer cells. Biochem Biophys Res Commun 430:1319–1321. https://doi.org/10.1016/j.bbrc.2012.12.031
Lefort CT, Wojciechowski K, Hocking DC (2011) N-cadherin cell-cell adhesion complexes are regulated by fibronectin matrix assembly. J Biol Chem 286:3149–3160. https://doi.org/10.1074/jbc.M110.115733
Liu X, Meng L, Li X, Li D, Liu Q, Chen Y, Li X, Bu W, Sun H (2020) Regulation of FN1 degradation by the p62/SQSTM1-dependent autophagy-lysosome pathway in HNSCC. Int J Oral Sci 12:34. https://doi.org/10.1038/s41368-020-00101-5
Abduljauwad SN, Ahmed HU (2019) Enhancing cancer cell adhesion with clay nanoparticles for countering metastasis. Sci Rep 9:5935
Hellinger JW, Schömel F, Buse JV, Lenz C, Bauerschmitz G, Emons G et al (2020) Identification of drivers of breast cancer invasion by secretome analysis: insight into CTGF signaling. Sci Rep 10:17889
Buchsbaum RJ, Oh SY (2016) Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers (Basel) 8:19
Emon B, Bauer J, Jain Y, Jung B, Saif T (2018) Biophysics of tumor microenvironment and cancer metastasis: a mini review. Comput Struct Biotechnol J 16:279–287
Butcher DT, Alliston T, Weaver VM (2009) A tense situation: forcing tumour progression. Nat Rev Cancer 9:108–122
Sullivan LB, Luengo A, Danai LV, Bush LN, Diehl FF, Hosios AM et al (2018) Aspartate is an endogenous metabolic limitation for tumour growth. Nat Cell Biol 20:782–788
Tong Y, Gao WQ, Liu Y (2020) Metabolic heterogeneity in cancer: an overview and therapeutic implications. Biochim Biophys Acta Rev Cancer 1874:188421
Kondo H, Ratcliffe C, Hooper S, Ellis J, MacRae JI, Hennequart M et al (2021) Single-cell resolved imaging reveals intra-tumor heterogeneity in glycolysis, transitions between metabolic states, and their regulatory mechanisms. Cell Rep 34:108750
Sarmasti Emami S, Zhang D, Yang X (2020) Interaction of the Hippo pathway and phosphatases in tumorigenesis. Cancers (Basel) 12:2438. https://doi.org/10.3390/cancers12092438
Strepkos D, Markouli M, Papavassiliou KA, Papavassiliou AG, Piperi C (2022) Emerging roles for the YAP/TAZ transcriptional regulators in brain tumour pathology and targeting options. Neuropathol Appl Neurobiol 48:e12762. https://doi.org/10.1111/nan.12762
Kim NG, Gumbiner BM (2015) Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol 210:503–515. https://doi.org/10.1083/jcb.201501025
Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12:31–46
Zhu Y, Zhu X, Tang C, Guan X, Zhang W (2021) Progress and challenges of immunotherapy in triple-negative breast cancer. Biochim Biophys Acta Rev Cancer 1876:188593
Geng QS, Huang T, Li LF, Shen ZB, Xue WH, Zhao J (2021) Over-expression and prognostic significance of fn1, correlating with immune infiltrates in thyroid cancer. Front Med (Lausanne) 8:812278
Sheng S, Guo B, Wang Z, Zhang Z, Zhou J, Huo Z (2021) Aberrant methylation and immune microenvironment are associated with overexpressed fibronectin 1: a diagnostic and prognostic target in head and neck squamous cell carcinoma. Front Mol Biosci 8:753563
Acknowledgements
None.
Funding
This study was supported by National Natural Science Foundation of China (82073146 and 82072903).
Author information
Authors and Affiliations
Contributions
CC and LY conceived the study, did the experiment, analyzed the data and writing the manuscript; JY analyzed and proofread the data; ZL and TL supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All experimental protocols were approved by the Animal Ethics Committee of Harbin Medical University Cancer Hospital. All methods were carried out in accordance with the Guide for the Care and Use of Laboratory Animals. All methods are reported in accordance with ARRIVE guidelines for the reporting of animal experiments. The study was carried out in accordance with the relevant guidelines and regulation.
Informed consent
Informed consent was obtained from all subjects prior to collection and all samples were subjected to histological confirmation. The study was conducted in accordance with the Declaration of Helsinki.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
In the original publication of the article, the name and email address of the first corresponding author was published incorrectly as Tong Liu, liutong@hrbmu.edu.cn. The corrected name should read as Tang Liu and the corrected email address should read as liutang0808@163.com. Also, the layout ratio was incorrect in Fig. 4. The corrected Fig. 4 is provided below.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, C., Ye, L., Yi, J. et al. FN1 mediated activation of aspartate metabolism promotes the progression of triple-negative and luminal a breast cancer. Breast Cancer Res Treat 201, 515–533 (2023). https://doi.org/10.1007/s10549-023-07032-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-07032-9