Abstract
Purpose
To determine the genetic and immune features associated with the recurrence of human epidermal growth factor receptor2-positive (HER2 +) breast cancer (BC) after trastuzumab-based treatment.
Methods
A retrospective cohort study of 48 patients who received trastuzumab-based treatment was divided into recurrent and non-recurrent groups according to clinical follow-up. Baseline samples from all 48 patients were analyzed for genetic variation, HLA allele type, gene expression, and immune features, which were linked to HER2 + BC recurrence. Statistics included logistic regression models, Kaplan–Meier plots, and Univariate Cox proportional hazards models.
Results
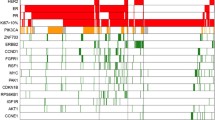
Compared with the non-recurrent group, the extracellular matrix-related pathway and 3 Hallmark gene sets were enriched in the recurrent group. The infiltration levels of immature B cells and activated B cells were significantly increased in the non-recurrent group, which correlated remarkably with improved overall survival (OS) in two other published gene expression datasets, including TCGA and METABRIC. In the TCGA cohort (n = 275), activated B cells (HR 0.23, 95%CI 0.13–0.43, p < 0.0001), and immature B cells (HR 0.26, 95%CI 0.12–0.59, p < 0.0001). In the METABRIC cohort (n = 236), activated B cells (HR 0.60, 95%CI 0.43–0.83, p = 0.002), and immature B cells (HR 0.65, 95%CI 0.47–0.91, p = 0.011). Cox regression suggested that immature B cells and activated B cells were protective factors for outcome OS.
Conclusions
Aberrant activation of multiple pathways and low baseline tumor-infiltrating B cells are related to HER2 + BC trastuzumab-based recurrence, which primarily affects the antitumor activity of trastuzumab.







Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- BC:
-
Breast cancer
- HER2 + :
-
HER2-positive
- TILs:
-
Tumor-infiltrating lymphocytes
- FFPE:
-
Formalin-fixed, paraffin-embedded
- SNV:
-
Single-nucleotide variants
- CNAs:
-
Copy number alterations
- ExAC:
-
Exome Aggregation Consortium
- gnomAD:
-
Genome Aggregation Database
- CNV:
-
Copy number variation
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
Gene ontology
- BP:
-
Biological process,
- CC:
-
Cellular component
- MF:
-
Molecular function
- ECM:
-
Extracellular matrix
- OS:
-
Overall survival
- ADCC:
-
Antibody-dependent cytotoxic cells
- IDC:
-
Invasive ductal carcinoma
- FAK1 :
-
Focal Adhesion Kinase
- RTK:
-
Receptor tyrosine kinases
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 321:288–300. https://doi.org/10.1001/jama.2018.19323
Cataldo A, Piovan C, Plantamura I, D’Ippolito E, Camelliti S, Casalini P, Giussani M, Deas O, Cairo S, Judde JG, Tagliabue E, Iorio MV (2018) MiR-205 as a predictive biomarker and adjuvant therapeutic tool in combination with trastuzumab. Oncotarget 9:27920–27928. https://doi.org/10.18632/oncotarget.24723
Rimawi M, De Angelis C, Contreras A, Pareja F, Geyer F, Burke K, Herrera S, Wang T, Mayer I, Forero A, Nanda R, Goetz M, Chang J, Krop I, Wolff A, Pavlick A, Fuqua S, Gutierrez C, Hilsenbeck S, Li M, Weigelt B, Reis-Filho J, Kent Osborne C, Schiff R (2018) Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer. Breast Cancer Res Treat 167:731–740. https://doi.org/10.1007/s10549-017-4533-9
Choi J, Jeon C, Kim Y, Jung S (2020) Pathological complete response to neoadjuvant trastuzumab and pertuzumab therapy is related to human epidermal growth factor receptor 2 (HER2) amplification level in HER2-amplified breast cancer. Medicine 99:e23053. https://doi.org/10.1097/md.0000000000023053
Yonemori K, Tsuta K, Shimizu C, Hatanaka Y, Hirakawa A, Ono M, Kouno T, Katsumata N, Ando M, Tamura K, Hasegawa T, Kinoshita T, Fujiwara Y (2010) Immunohistochemical expression of HER1, HER3, and HER4 in HER2-positive breast cancer patients treated with trastuzumab-containing neoadjuvant chemotherapy. J Surg Oncol 101:222–227. https://doi.org/10.1002/jso.21486
Denkert C, Huober J, Loibl S, Prinzler J, Kronenwett R, Darb-Esfahani S, Brase J, Solbach C, Mehta K, Fasching P, Sinn B, Engels K, Reinisch M, Hansmann M, Tesch H, von Minckwitz G, Untch M (2013) HER2 and ESR1 mRNA expression levels and response to neoadjuvant trastuzumab plus chemotherapy in patients with primary breast cancer. Breast Cancer Res 15:R11. https://doi.org/10.1186/bcr3384
Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, Piccart MJ, Loibl S, Denkert C, Smyth MJ, Joensuu H, Sotiriou C (2014) Tumor-infiltrating lymphocytes are prognostic in triple-negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol 25:1544–1550. https://doi.org/10.1093/annonc/mdu112
Perez E, Thompson E, Ballman K, Anderson S, Asmann Y, Kalari K, Eckel-Passow J, Dueck A, Tenner K, Jen J, Fan J, Geiger X, McCullough A, Chen B, Jenkins R, Sledge G, Winer E, Gralow J, Reinholz M (2015) Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the north central cancer treatment group n9831 adjuvant trastuzumab trial. J Clin Oncol 33:701–708. https://doi.org/10.1200/jco.2014.57.6298
Varadan V, Gilmore H, Miskimen K, Tuck D, Parsai S, Awadallah A, Krop I, Winer E, Bossuyt V, Somlo G, Abu-Khalaf M, Fenton M, Sikov W, Harris L (2016) Immune signatures following single dose trastuzumab predict pathologic response to preoperativetrastuzumab and chemotherapy in HER2-positive early breast cancer. Clin Cancer Res 22:3249–3259. https://doi.org/10.1158/1078-0432.ccr-15-2021
Hou Y, Nitta H, Wei L, Banks P, Parwani A, Li Z (2018) Evaluation of immune reaction and PD-L1 expression using multiplex immunohistochemistry in HER2-positive breast cancer: the association with response to anti-HER2 neoadjuvant therapy. Clin Breast Cancer 18:e237–e244. https://doi.org/10.1016/j.clbc.2017.11.001
Li H (2013) Aligning sequence reads, clone sequences, and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. https://doi.org/10.48550/arXiv.1303.3997
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. https://doi.org/10.1038/ng.806
Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC, Dry JR (2016) VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 44:e108. https://doi.org/10.1093/nar/gkw227
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. https://doi.org/10.48550/arXiv.1207.3907
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, Birnbaum D, The Exome Aggregation C, Daly MJ, MacArthur DG (2017) The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 45:D840–D845. https://doi.org/10.1093/nar/gkw971
Talevich E, Shain AH, Botton T, Bastian BC (2016) CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 12:e1004873
Kawaguchi S, Higasa K, Shimizu M, Yamada R, Matsuda F (2017) HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum Mutat 38:788–797. https://doi.org/10.1002/humu.23230
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. https://doi.org/10.1073/pnas.0506580102
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262. https://doi.org/10.1016/j.celrep.2016.12.019
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS (2016) Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165:35–44. https://doi.org/10.1016/j.cell.2016.02.065
Diermeier S, Horvath G, Knuechel-Clarke R, Hofstaedter F, Szollosi J, Brockhoff G (2005) Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp Cell Res 304:604–619. https://doi.org/10.1016/j.yexcr.2004.12.008
Maadi H, Nami B, Tong J, Li G, Wang Z (2018) The effects of trastuzumab on HER2-mediated cell signaling in CHO cells expressing human HER2. BMC Cancer 18:238. https://doi.org/10.1186/s12885-018-4143-x
Guo P, Pu T, Chen S, Qiu Y, Zhong X, Zheng H, Chen L, Bu H, Ye F (2017) Breast cancers with EGFR and HER2 co-amplification favor distant metastasis and poor clinical outcome. Oncol Lett 14:6562–6570. https://doi.org/10.3892/ol.2017.7051
Cheng H, Ballman K, Vassilakopoulou M, Dueck AC, Reinholz MM, Tenner K, Gralow J, Hudis C, Davidson NE, Fountzilas G, McCullough AE, Chen B, Psyrri A, Rimm DL, Perez EA (2014) EGFR expression is associated with decreased benefit from trastuzumab in the NCCTG N9831 (Alliance) trial. Br J Cancer 111:1065–1071. https://doi.org/10.1038/bjc.2014.442
Toth G, Szoor A, Simon L, Yarden Y, Szollosi J, Vereb G (2016) The combination of trastuzumab and pertuzumab administered at approved doses may delay development of trastuzumab resistance by additively enhancing antibody-dependent cell-mediated cytotoxicity. MAbs 8:1361–1370. https://doi.org/10.1080/19420862.2016.1204503
Dillon PM, Brenin CM, Slingluff CL Jr (2020) Evaluating Nelipepimut-S in the treatment of breast cancer: a short report on the emerging data. Breast Cancer 12:69–75. https://doi.org/10.2147/bctt.s224758
McCarthy P, Clifton G, Vreeland T, Adams A, O’Shea A, Peoples G (2021) AE37: a HER2-targeted vaccine for the prevention of breast cancer recurrence. Expert Opin Investig Drugs 30:5–11. https://doi.org/10.1080/13543784.2021.1849140
Gamzatova Z, Villabona L, Dahlgren L, Dalianis T, Nillson B, Bergfeldt K, Masucci GV (2006) Human leucocyte antigen (HLA) A2 as a negative clinical prognostic factor in patients with advanced ovarian cancer. Gynecol Oncol 103:145–150. https://doi.org/10.1016/j.ygyno.2006.02.004
Andersson E, Villabona L, Bergfeldt K, Carlson JW, Ferrone S, Kiessling R, Seliger B, Masucci GV (2012) Correlation of HLA-A02* genotype and HLA class I antigen down-regulation with the prognosis of epithelial ovarian cancer. Cancer Immunol Immunother 61:1243–1253. https://doi.org/10.1007/s00262-012-1201-0
Andersson E, Poschke I, Villabona L, Carlson JW, Lundqvist A, Kiessling R, Seliger B, Masucci GV (2016) Non-classical HLA-class I expression in serous ovarian carcinoma: correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology 5:e1052213. https://doi.org/10.1080/2162402X.2015.1052213
Mittendorf EA, Lu B, Melisko M, Price Hiller J, Bondarenko I, Brunt AM, Sergii G, Petrakova K, Peoples GE (2019) Efficacy and safety analysis of Nelipepimut-S vaccine to prevent breast cancer recurrence: a randomized, multicenter, phase III clinical trial. Clin Cancer Res 25:4248–4254. https://doi.org/10.1158/1078-0432.CCR-18-2867
Insua-Rodriguez J, Oskarsson T (2016) The extracellular matrix in breast cancer. Adv Drug Deliv Rev 97:41–55. https://doi.org/10.1016/j.addr.2015.12.017
Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ (2010) HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat 122:35–43. https://doi.org/10.1007/s10549-009-0502-2
Jena MK, Janjanam J (2018) Role of extracellular matrix in breast cancer development: a brief update. F1000Res 7:274. https://doi.org/10.12688/f1000research.14133.2
Im J, Fu W, Wang H, Bhatia S, Hammer D, Kowalska M, Muschel R (2004) Coagulation facilitates tumor cell spreading in the pulmonary vasculature during early metastatic colony formation. Cancer Res 64:8613–8619. https://doi.org/10.1158/0008-5472.can-04-2078
Li J, Du J, Wang Y, Jia H (2021) A coagulation-related gene-based prognostic model for invasive ductal carcinoma. Front Genet 12:722992. https://doi.org/10.3389/fgene.2021.722992
Sulzmaier FJ, Jean C, Schlaepfer DD (2014) FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer 14:598–610. https://doi.org/10.1038/nrc3792
Martin Schwill RT, Gajadhar Aaron S, Kast Florian, White Forest M, Plückthun Andreas (2019) Systemic analysis of tyrosine kinase signaling reveals a common adaptive response program in a HER2-positive breast cancer. Sci Signal 12:2875. https://doi.org/10.1126/scisignal.aau2875
Guarneri V, Frassoldati A, Bottini A, Cagossi K, Bisagni G, Sarti S, Ravaioli A, Cavanna L, Giardina G, Musolino A, Untch M, Orlando L, Artioli F, Boni C, Generali DG, Serra P, Bagnalasta M, Marini L, Piacentini F, D’Amico R, Conte P (2012) Preoperative chemotherapy plus trastuzumab, lapatinib, or both in human epidermal growth factor receptor 2-positive operable breast cancer: results of the randomized phase II CHER-LOB study. J Clin Oncol 30:1989–1995. https://doi.org/10.1200/JCO.2011.39.0823
de Azambuja E, Holmes AP, Piccart-Gebhart M, Holmes E, Di Cosimo S, Swaby RF, Untch M, Jackisch C, Lang I, Smith I, Boyle F, Xu B, Barrios CH, Perez EA, Azim HA Jr, Kim SB, Kuemmel S, Huang CS, Vuylsteke P, Hsieh RK, Gorbunova V, Eniu A, Dreosti L, Tavartkiladze N, Gelber RD, Eidtmann H, Baselga J (2014) Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): survival outcomes of a randomized, open-label, multicentre, phase 3 trial and their association with pathological complete response. Lancet Oncol 15:1137–1146. https://doi.org/10.1016/S1470-2045(14)70320-1
Perez E, Ballman K, Tenner K, Thompson E, Badve S, Bailey H, Baehner F (2016) Association of stromal tumor-infiltrating lymphocytes with recurrence-free survival in the N9831 adjuvant trial in patients with early-stage HER2-positive breast cancer. JAMA Oncol 2:56–64. https://doi.org/10.1001/jamaoncol.2015.3239
Chumsri S, Li Z, Serie D, Norton N, Mashadi-Hossein A, Tenner K, Brauer H, Warren S, Danaher P, Colon-Otero G, Partridge A, Carey L, Hilbers F, Van Dooren V, Holmes E, Di Cosimo S, Werner O, Huober J, Dueck A, Sotiriou C, Saura C, Moreno-Aspitia A, Knutson K, Perez E, Thompson E (2022) Adaptive immune signature in HER2-positive breast cancer in NCCTG (Alliance) N9831 and NeoALTTO trials. NPJ Breast Cancer 8:68. https://doi.org/10.1038/s41523-022-00430-0
Gu X, Zhang Q, Wu X, Fan Y, Qian J (2021) Gene coexpression network approach to develop an immune prognostic model for pancreatic adenocarcinoma. World J Surg Oncol 19:112. https://doi.org/10.1186/s12957-021-02201-w
Acknowledgements
Not applicable.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
CX and YW conceived and designed the study. YH and RY supervised and administrated the project. LW, XS, YQ, and ZZ conducted the experiments, and WZ, YY, and WC acquired and analyzed the data. YZ and ZL drafted the manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The study was conducted with the approval of the Medical Ethics Committees of the Peking Union Medical College Hospital.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2023_6931_MOESM2_ESM.tif
Supplementary file2 (TIF 10910 KB)—Fig. S2 GSEA of Hallmark gene sets in recurrent and non-recurrent HER2+ BC patients. Significantly enriched signaling pathways were revealed by gene set enrichment analysis. At the top of the plot is the run enrichment score (ES) for the gene set, and the score at the peak (the score furthest from 0.0) represents the ES for the entire gene set. The middle section shows where the members of the genome appear in the sorted list of genes. The bottom portion of the plot shows that the value of the ranking metric decreases as the value of the ranking gene list reduces
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, C., Wang, Y., Hong, Y. et al. Identification of genetic and immune signatures for the recurrence of HER2-positive breast cancer after trastuzumab-based treatment. Breast Cancer Res Treat 199, 603–615 (2023). https://doi.org/10.1007/s10549-023-06931-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06931-1