Abstract
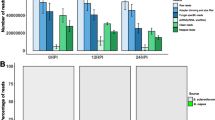
Non-coding, small RNAs (sRNAs) have been identified in a wide spectrum of organisms ranging from bacteria to humans; however, the role and mechanisms of these sRNA in plant immunity is largely unknown. To determine possible roles of sRNA in plant–pathogen interaction, we carried out a high-throughput sRNA sequencing of Brassica campestris using non-infected plants and plants infected with Erwinia carotovora. Consistent with our hypothesis that distinct classes of host sRNAs alerts their expression levels in response to infection, we found that: (1) host 28-nt sRNAs were strongly increased under pathogen infection; and (2) a group of host sRNAs homologous to the pathogen genome also accumulated at significantly higher level. Our data thus suggest several distinct classes of the host sRNAs may enhance their function by up-regulation of their expression/stability in response to bacterial pathogen challenges.







Similar content being viewed by others
References
Aguilar I, Alamillo JM, Garcia-Olmedo F, Rodriguez-Palenzuela P (2002) Natural variability in the Arabidopsis response to infection with Erwinia carotovora subsp. carotovora. Planta 215:205–209
Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673–676
Anderson MT, Seifert HS (2011) Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen. mBio 2:e00005–e00011
Bell KS, Avrova AO, Holeva MC, Cardle L et al (2002) Sample sequencing of a selected region of the genome of Erwinia carotovora subsp. atroseptica reveals candidate phytopathogenicity genes and allows comparison with Escherichia coli. Microbiology 148:1367–1378
Boyle R (2011) Fragment of human DNA found in genome of gonorrhea bacteria. POPSCI Publishing PhysicsWebhttp://www.popsci.com/science/article/2011-02/v-day-study-shows-human-dna-fragment-genome-common-vd-bacteria
Chen HM, Li YH, Wu SH (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA 104:3318–3323
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Dyall SD, Brown MT, Johnson PJ (2004) Ancient invasions: from endosymbionts to organelles. Science 304:253–257
Ellendorff U, Fradin EF, de Jonge R, Thomma BP (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot 60:591–602
Guo L, Lu Z (2010) The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE 5:e11387
He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582:2445–2452
Hu Q, Hollunder J, Niehl A, Korner CJ et al (2011) Specific impact of tobamovirus infection on the Arabidopsis small RNA profile. PLoS ONE 6:e19549
Jin H (2008) Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett 582:2679–2684
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr, Zhu JK, Staskawicz BJ, Jin H (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103:18002–18007
Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21:3123–3134
Kozjak-Pavlovic V, Dian-Lothrop EA, Meinecke M, Kepp O et al (2009) Bacterial porin disrupts mitochondrial membrane potential and sensitizes host cells to apoptosis. PLoS Pathog 5:e1000629
Kuchenbauer F, Mah SM, Heuser M, McPherson A et al (2011) Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 118:3350–3358
Kume K, Tsutsumi K, Saitoh Y (2010) TAS1 trans-acting siRNA targets are differentially regulated at low temperature, and TAS1 trans-acting siRNA mediates temperature-controlled At1g51670 expression. Biosci Biotechnol Biochem 74:1435–1440
Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152:2222–2231
Moldovan D, Spriggs A, Dennis ES, Wilson IW (2010) The hunt for hypoxia responsive natural antisense short interfering RNAs. Plant Signal Behav 5:247–251
Montgomery TA, Howell MD, Cuperus JT, Li D et al (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128–141
Mosher RA, Baulcombe DC (2008) Bacterial pathogens encode suppressors of RNA-mediated silencing. Genome Biol 9:237
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321:964–967
Ren J, Petzoldt R, Dickson MH (2001) Screening and identification of resistance to bacterial soft rot in Brassica rapa. Euphytica 118:271–280
Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyan J, Masuta C (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 7:e1002021
Smith NA, Eamens AL, Wang MB (2011) Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 7:e1002022
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Vanjildorj E, Song SY, Yang ZH, Choi JE et al (2009) Enhancement of tolerance to soft rot disease in the transgenic Chinese cabbage (Brassica rapa L. ssp. pekinensis) inbred line, Kenshin. Plant Cell Rep 28:1581–1591
Wang X, Wang H, Wang J, Sun R et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC (2011) Widespread regulatory activity of vertebrate microRNA* species. RNA 17:312–326
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE Gene MEMB12. Mol Cell 42:356–366
Acknowledgments
The author would like to thank Dr. Yizhen Lu and Xiang Yu for their computational assistance and helpful advice during manuscript production. This work was supported by a grant from the National Natural Science Foundation of China (No. 30871617) and a grant from the Science and Technology Commission of Shanghai Municipality (No. 12ZR1435600).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
The analysis of northern blot hybridization confirmed the accumulation level of the sRNAs from the control or the Ecc-inoculated samples was consistent with deep-sequencing results. Mock: the sRNAs from the control samples; Ecc-infection: the sRNAs from the Ecc-inoculated samples. miRNA156a, miRNA159a, miRNA319a, and miRNA398b are selected for northern blot hybridization analysis, the ubiquitin RNA hybridization is used as a loading control (TIFF 500 kb)
Rights and permissions
About this article
Cite this article
Sun, C.B. Characterization of the small RNA transcriptome in plant–microbe (Brassica/Erwinia) interactions by high-throughput sequencing. Biotechnol Lett 36, 371–381 (2014). https://doi.org/10.1007/s10529-013-1362-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1362-8