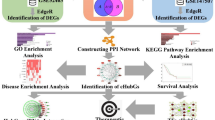
Abstract
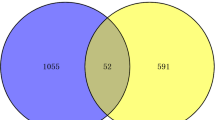
The number of patients with COVID-19 caused by severe acute respiratory syndrome coronavirus 2 is still increasing. In the case of COVID-19 and tuberculosis (TB), the presence of one disease affects the infectious status of the other. Meanwhile, coinfection may result in complications that make treatment more difficult. However, the molecular mechanisms underpinning the interaction between TB and COVID-19 are unclear. Accordingly, transcriptome analysis was used to detect the shared pathways and molecular biomarkers in TB and COVID-19, allowing us to determine the complex relationship between COVID-19 and TB. Two RNA-seq datasets (GSE114192 and GSE163151) from the Gene Expression Omnibus were used to find concerted differentially expressed genes (DEGs) between TB and COVID-19 to identify the common pathogenic mechanisms. A total of 124 common DEGs were detected and used to find shared pathways and drug targets. Several enterprising bioinformatics tools were applied to perform pathway analysis, enrichment analysis and networks analysis. Protein–protein interaction analysis and machine learning was used to identify hub genes (GAS6, OAS3 and PDCD1LG2) and datasets GSE171110, GSE54992 and GSE79362 were used for verification. The mechanism of protein-drug interactions may have reference value in the treatment of coinfection of COVID-19 and TB.












Similar content being viewed by others
Data Availability
Data sharing not applicable—no new data generated.
References
Anderson G, Carbone A, Mazzoccoli G (2020) Aryl hydrocarbon receptor role in co-ordinating SARS-CoV-2 entry and symptomatology: linking cytotoxicity changes in COVID-19 and cancers modulation by racial discrimination stress. Biology. https://doi.org/10.3390/biology9090249
Anderson G, Carbone A, Mazzoccoli G (2021) Tryptophan metabolites and aryl hydrocarbon receptor in severe acute respiratory syndrome, coronavirus-2 (SARS-CoV-2) pathophysiology. Int J Mol Sci. https://doi.org/10.3390/ijms22041597
Antas PR et al (2006) Decreased CD4+ lymphocytes and innate immune responses in adults with previous extrapulmonary tuberculosis. J Allergy Clin Immunol 117(4):916–923. https://doi.org/10.1016/j.jaci.2006.01.042
Artese A et al (2020) Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses. Drug Resist Updat 53:100721. https://doi.org/10.1016/j.drup.2020.100721
Ashburner M et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Attia EF et al (2019) Tuberculosis and other bacterial co-infection in Cambodia: a single center retrospective cross-sectional study. BMC Pulm Med 19(1):60. https://doi.org/10.1186/s12890-019-0828-4
Barrett T et al (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. https://doi.org/10.1093/nar/gks1193
Barst RJ et al (2002) Clinical efficacy of sitaxsentan, an endothelin-A receptor antagonist, in patients with pulmonary arterial hypertension: open-label pilot study. Chest 121(6):1860–1868. https://doi.org/10.1378/chest.121.6.1860
Belenky P, Bogan KL, Brenner C (2007) NAD+ metabolism in health and disease. Trends Biochem Sci 32(1):12–19. https://doi.org/10.1016/j.tibs.2006.11.006
Chai Q, Zhang Y, Liu CH (2018) Mycobacterium tuberculosis: an adaptable pathogen associated with multiple human diseases. Front Cell Infect Microbiol 8:158. https://doi.org/10.3389/fcimb.2018.00158
Chang WT et al (2021) Cardiac involvement of COVID-19: a comprehensive review. Am J Med Sci 361(1):14–22. https://doi.org/10.1016/j.amjms.2020.10.002
Chin C-H et al (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(S4):S11. https://doi.org/10.1186/1752-0509-8-s4-s11
Cho Y et al (2020) Identification of serum biomarkers for active pulmonary tuberculosis using a targeted metabolomics approach. Sci Rep 10(1):3825. https://doi.org/10.1038/s41598-020-60669-0
Crisan-Dabija R et al (2020) Tuberculosis and COVID-19: lessons from the past viral outbreaks and possible future outcomes. Can Respir J 2020:1401053. https://doi.org/10.1155/2020/1401053
Crowther RR, Qualls JE (2020) Metabolic regulation of immune responses to mycobacterium tuberculosis: A spotlight on l-arginine and l-tryptophan metabolism. Front Immunol 11:628432. https://doi.org/10.3389/fimmu.2020.628432
da Huang W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. https://doi.org/10.1093/nar/gkn923
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
De Flora S, Balansky R, La Maestra S (2020) Rationale for the use of N-acetylcysteine in both prevention and adjuvant therapy of COVID-19. Faseb j 34(10):13185–13193. https://doi.org/10.1096/fj.202001807
Diao B et al (2020) Reduction and functional exhaustion of t cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 11:827. https://doi.org/10.3389/fimmu.2020.00827
Fan J et al (2018) MiR-665 aggravates heart failure via suppressing CD34-mediated coronary microvessel angiogenesis. Aging 10(9):2459–2479. https://doi.org/10.18632/aging.101562
Feng S et al (2015) Analysis of serum metabolic profile by ultra-performance liquid chromatography-mass spectrometry for biomarkers discovery: application in a pilot study to discriminate patients with tuberculosis. Chin Med J 128(2):159–168. https://doi.org/10.4103/0366-6999.149188
Fenizia C et al (2021) SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells. https://doi.org/10.3390/cells10061434
Feruglio SL et al (2015) Early dynamics of T helper cell cytokines and T regulatory cells in response to treatment of active Mycobacterium tuberculosis infection. Clin Exp Immunol 179(3):454–465. https://doi.org/10.1111/cei.12468
Fogel N (2015) Tuberculosis: a disease without boundaries. Tuberculosis 95(5):527–531. https://doi.org/10.1016/j.tube.2015.05.017
Fornes O et al (2020) JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48(D1):D87-d92. https://doi.org/10.1093/nar/gkz1001
Franco-Paredes C et al (2018) Cutaneous mycobacterial infections. Clin Microbiol Rev. https://doi.org/10.1128/cmr.00069-18
Fu X et al (2018) Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 128(5):2127–2143. https://doi.org/10.1172/jci98215
Geng L, Liu W, Chen Y (2017) miR-124-3p attenuates MPP(+)-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp Biol Med (maywood) 242(18):1757–1764. https://doi.org/10.1177/1535370217734492
Hays E, Bonavida B (2019) YY1 regulates cancer cell immune resistance by modulating PD-L1 expression. Drug Resist Updat 43:10–28. https://doi.org/10.1016/j.drup.2019.04.001
Heitmann L et al (2014) The IL-13/IL-4Rα axis is involved in tuberculosis-associated pathology. J Pathol 234(3):338–350. https://doi.org/10.1002/path.4399
Hsu SD et al (2011) miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq1107
Huang KC et al (2020) Prognostic relevance of programmed cell death 1 ligand 2 (PDCD1LG2/PD-L2) in patients with advanced stage colon carcinoma treated with chemotherapy. Sci Rep 10(1):22330. https://doi.org/10.1038/s41598-020-79419-3
Isa F et al (2018) Mass spectrometric identification of urinary biomarkers of pulmonary tuberculosis. EBioMedicine 31:157–165. https://doi.org/10.1016/j.ebiom.2018.04.014
Iwata-Yoshikawa N et al (2019) TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. https://doi.org/10.1128/jvi.01815-18
Jiang F et al (2021) LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by Mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. Mol Immunol 139:42–49. https://doi.org/10.1016/j.molimm.2021.07.023
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Lambert SA et al (2018) The human transcription factors. Cell 172(4):650–665. https://doi.org/10.1016/j.cell.2018.01.029
Larrouy-Maumus G (2019) Lipids as biomarkers of cancer and bacterial infections. Curr Med Chem 26(11):1924–1932. https://doi.org/10.2174/0929867325666180904120029
Li G, Zhang L (2021) miR-335-5p aggravates type 2 diabetes by inhibiting SLC2A4 expression. Biochem Biophys Res Commun 558:71–78. https://doi.org/10.1016/j.bbrc.2021.04.011
Lv L et al (2021) MiR-124-3p reduces angiotensin II-dependent hypertension by down-regulating EGR1. J Hum Hypertens 35(8):696–708. https://doi.org/10.1038/s41371-020-0381-x
Mandal N et al (2020) Correlation between CNS tuberculosis and the COVID-19 pandemic: the neurological and therapeutic insights. ACS Chem Neurosci 11(18):2789–2792. https://doi.org/10.1021/acschemneuro.0c00546
McGonagle D, Ramanan AV, Bridgewood C (2021) Immune cartography of macrophage activation syndrome in the COVID-19 era. Nat Rev Rheumatol 17(3):145–157. https://doi.org/10.1038/s41584-020-00571-1
Meo SA et al (2021) Omicron SARS-CoV-2 new variant: global prevalence and biological and clinical characteristics. Eur Rev Med Pharmacol Sci 25(24):8012–8018. https://doi.org/10.26355/eurrev_202112_27652
Miotto D et al (2001) Expression of IFN-gamma-inducible protein; monocyte chemotactic proteins 1, 3, and 4; and eotaxin in TH1- and TH2-mediated lung diseases. J Allergy Clin Immunol 107(4):664–670. https://doi.org/10.1067/mai.2001.113524
Mirzaei R et al (2020) Bacterial co-infections with SARS-CoV-2. IUBMB Life 72(10):2097–2111. https://doi.org/10.1002/iub.2356
Miyashita N et al (2020) FOXL1 regulates lung fibroblast function via multiple mechanisms. Am J Respir Cell Mol Biol 63(6):831–842. https://doi.org/10.1165/rcmb.2019-0396OC
Morrison VA (2014) Immunosuppression associated with novel chemotherapy agents and monoclonal antibodies. Clin Infect Dis 59(Suppl 5):S360–S364. https://doi.org/10.1093/cid/ciu592
Motta I et al (2020) Tuberculosis, COVID-19 and migrants: preliminary analysis of deaths occurring in 69 patients from two cohorts. Pulmonology 26(4):233–240. https://doi.org/10.1016/j.pulmoe.2020.05.002
Mousquer GT, Peres A, Fiegenbaum M (2021) Pathology of TB/COVID-19 co-infection: the phantom menace. Tuberculosis 126:102020. https://doi.org/10.1016/j.tube.2020.102020
Muefong CN, Sutherland JS (2020) Neutrophils in tuberculosis-associated inflammation and lung pathology. Front Immunol 11:962. https://doi.org/10.3389/fimmu.2020.00962
Petruk G et al (2020) SARS-CoV-2 spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J Mol Cell Biol 12(12):916–932. https://doi.org/10.1093/jmcb/mjaa067
Polidoro RB et al (2020) Overview: systemic inflammatory response derived from lung injury caused by SARS-CoV-2 infection explains severe outcomes in COVID-19. Front Immunol 11:1626. https://doi.org/10.3389/fimmu.2020.01626
Prieto-Peña D, Dasgupta B (2021) Biologic agents and small-molecule inhibitors in systemic autoimmune conditions: an update. Pol Arch Intern Med 131(2):171–181. https://doi.org/10.20452/pamw.15438
Puk O et al (2022) Pulmonary artery targeted therapy in treatment of COVID-19 related ARDS. Lit Rev Biomed Pharmacother 146:112592. https://doi.org/10.1016/j.biopha.2021.112592
Rahman MH et al (2020) A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph17031035
Sahler J, Woeller CF, Phipps RP (2014) Microparticles engineered to highly express peroxisome proliferator-activated receptor-γ decreased inflammatory mediator production and increased adhesion of recipient monocytes. PLoS ONE 9(11):e113189. https://doi.org/10.1371/journal.pone.0113189
Samudrala PK et al (2020) Virology, pathogenesis, diagnosis and in-line treatment of COVID-19. Eur J Pharmacol 883:173375. https://doi.org/10.1016/j.ejphar.2020.173375
Schmidt AF et al (2021) Cholesteryl ester transfer protein (CETP) as a drug target for cardiovascular disease. Nat Commun 12(1):5640. https://doi.org/10.1038/s41467-021-25703-3
Schriml LM et al (2019) Human disease ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 47(D1):D955-d962. https://doi.org/10.1093/nar/gky1032
Shen C et al (2020) Advancement of E2F1 in common tumors. Zhongguo Fei Ai Za Zhi 23(10):921–926. https://doi.org/10.3779/j.issn.1009-3419.2020.101.32
Shi YP et al (2020) miR-17-5p knockdown inhibits proliferation, autophagy and promotes apoptosis in thyroid cancer via targeting PTEN. Neoplasma 67(2):249–258. https://doi.org/10.4149/neo_2019_190110N29
Sidharta PN, Treiber A, Dingemanse J (2015) Clinical pharmacokinetics and pharmacodynamics of the endothelin receptor antagonist macitentan. Clin Pharmacokinet 54(5):457–471. https://doi.org/10.1007/s40262-015-0255-5
Spinner MA et al (2014) GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood 123(6):809–821. https://doi.org/10.1182/blood-2013-07-515528
Srirojnopkun C et al (2018) Association of APOE and CETP TaqIB polymorphisms with type 2 diabetes mellitus. Arch Med Res 49(7):479–485. https://doi.org/10.1016/j.arcmed.2019.02.005
Stochino C et al (2020) Clinical characteristics of COVID-19 and active tuberculosis co-infection in an Italian reference hospital. Eur Respir J. https://doi.org/10.1183/13993003.01708-2020
Sun D et al (2021) Overexpressed miR-335-5p reduces atherosclerotic vulnerable plaque formation in acute coronary syndrome. J Clin Lab Anal 35(2):e23608. https://doi.org/10.1002/jcla.23608
Szklarczyk D et al (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362–D368. https://doi.org/10.1093/nar/gkw937
Tapela K, Ochieng’ Olwal C, Quaye O (2020) Parallels in the pathogenesis of SARS-CoV-2 and M. tuberculosis: a synergistic or antagonistic alliance? Future Microbiol 15:1691–1695. https://doi.org/10.2217/fmb-2020-0179
Taus F et al (2020) Platelets promote thromboinflammation in SARS-CoV-2 pneumonia. Arterioscler Thromb Vasc Biol 40(12):2975–2989. https://doi.org/10.1161/atvbaha.120.315175
Tcheng JE et al (2003) Benefits and risks of abciximab use in primary angioplasty for acute myocardial infarction: the controlled abciximab and device investigation to lower late angioplasty complications (CADILLAC) trial. Circulation 108(11):1316–1323. https://doi.org/10.1161/01.Cir.0000087601.45803.86
Tumpara S et al (2021) Boosted pro-inflammatory activity in human PBMCs by lipopolysaccharide and SARS-CoV-2 spike protein is regulated by α-1 antitrypsin. Int J Mol Sci. https://doi.org/10.3390/ijms22157941
Tutusaus A et al (2020) Role of vitamin K-dependent factors protein S and GAS6 and TAM receptors in SARS-CoV-2 infection and COVID-19-associated immunothrombosis. Cells. https://doi.org/10.3390/cells9102186
Urban A et al (2021) Gain-of-function mutations R249C and S250C in complement C2 protein increase C3 deposition in the presence of C-reactive protein. Front Immunol 12:724361. https://doi.org/10.3389/fimmu.2021.724361
van den Berg MMJ et al (2020) Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog Neurobiol 185:101732. https://doi.org/10.1016/j.pneurobio.2019.101732
Vaz de Paula CB et al (2020) IL-4/IL-13 remodeling pathway of COVID-19 lung injury. Sci Rep 10(1):18689. https://doi.org/10.1038/s41598-020-75659-5
Venitz J et al (2012) Clinical pharmacokinetics and drug-drug interactions of endothelin receptor antagonists in pulmonary arterial hypertension. J Clin Pharmacol 52(12):1784–1805. https://doi.org/10.1177/0091270011423662
Vodnar DC et al (2020) Coronavirus disease (COVID-19) caused by (SARS-CoV-2) infections: a real challenge for human gut microbiota. Front Cell Infect Microbiol 10:575559. https://doi.org/10.3389/fcimb.2020.575559
Vrieling F et al (2019) Plasma metabolomics in tuberculosis patients with and without concurrent type 2 diabetes at diagnosis and during antibiotic treatment. Sci Rep 9(1):18669. https://doi.org/10.1038/s41598-019-54983-5
Walls AC et al (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281-292.e6. https://doi.org/10.1016/j.cell.2020.02.058
Wang XX et al (2021) Histopathological features of multiorgan percutaneous tissue core biopsy in patients with COVID-19. J Clin Pathol 74(8):522–527. https://doi.org/10.1136/jclinpath-2020-206623
Weiner J 3rd et al (2012) Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE 7(7):e40221. https://doi.org/10.1371/journal.pone.0040221
Wishart DS et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46(D1):D1074–D1082. https://doi.org/10.1093/nar/gkx1037
Wong M et al (2008) TNFalpha blockade in human diseases: mechanisms and future directions. Clin Immunol 126(2):121–136. https://doi.org/10.1016/j.clim.2007.08.013
Xia X, Wang Y, Zheng J (2021) COVID-19 and Alzheimer’s disease: how one crisis worsens the other. Transl Neurodegener 10(1):15. https://doi.org/10.1186/s40035-021-00237-2
Xiao X et al (2019) Exome sequencing reveals a heterozygous OAS3 mutation in a Chinese family with juvenile-onset open-angle glaucoma. Invest Ophthalmol vis Sci 60(13):4277–4284. https://doi.org/10.1167/iovs.19-27545
Yagi K et al (2010) Ibudilast inhibits cerebral aneurysms by down-regulating inflammation-related molecules in the vascular wall of rats. Neurosurgery 66(3):551–559. https://doi.org/10.1227/01.Neu.0000365771.89576.77
Yang L et al (2018) Lectin microarray combined with mass spectrometry identifies haptoglobin-related protein (HPR) as a potential serologic biomarker for separating nonbacterial pneumonia from bacterial pneumonia in childhood. Proteomics Clin Appl 12(6):e1800030. https://doi.org/10.1002/prca.201800030
Zhang Z et al (2020) Gene correlation network analysis to identify regulatory factors in sepsis. J Transl Med 18(1):381. https://doi.org/10.1186/s12967-020-02561-z
Zhao XG et al (2019) miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis 10(7):479. https://doi.org/10.1038/s41419-019-1705-z
Zheng XW et al (2016) Subcutaneous tuberculosis formation during FOLFIRI and bevacizumab treatment: a case report. Int J Colorectal Dis 31(4):943–944. https://doi.org/10.1007/s00384-015-2368-6
Zhou G et al (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47(W1):W234-w241. https://doi.org/10.1093/nar/gkz240
Zhu Y et al (2021) Blocking SNHG14 antagonizes lipopolysaccharides-induced acute lung injury via SNHG14/miR-124-3p axis. J Surg Res 263:140–150. https://doi.org/10.1016/j.jss.2020.10.034
Acknowledgements
We sincerely acknowledge the GEO database for offering their platform and their contributions for uploading their valuable dataset.
Funding
Not applicable for that section.
Author information
Authors and Affiliations
Contributions
ZH, JK, JXL, and PC designed the study, acquired all datasets, analyzed the data, prepared the figure and table and wrote the main manuscript. JL and YH analyzed the data, collected the specimens and interpreted the data. QL, HC, NH, and TL supervised the project and revised the manuscript. XG: evaluated and guided the full text and granted final approval of the version to be submitted. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
No potential conflict of interest was reported by the author(s).
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, ZM., Kang, JQ., Chen, PZ. et al. Identifying the Interaction Between Tuberculosis and SARS-CoV-2 Infections via Bioinformatics Analysis and Machine Learning. Biochem Genet (2023). https://doi.org/10.1007/s10528-023-10563-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-023-10563-x