Abstract
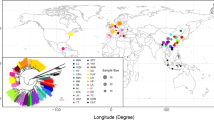
In this study, we identified porcine single nucleotide polymorphisms (SNPs) by aligning eight sequences generated with two approaches: amplification of 665 intronic regions using one sample from each of eight breeds, including three East Asian pigs, and amplification of 289 3′-UTR regions using two samples from each of four major commercial breeds. The 1,760 and 599 SNPs were validated using two 384-sample DNA panels by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. The phylogenetic tree and Structure analyses classified the pigs into two large clusters: Euro-American and East Asian populations. The membership proportions, however, differed between inferred clusters for K = 2 generated by the two approaches. With intronic SNPs, Euro-American breeds constituted about 100% of the Euro-American cluster, but with 3′-UTR SNPs, about 17% of the East Asian cluster comprised five Euro-American breeds. The differences in the SNP discovery panels may affect population structure found in study panels of large samples.


Similar content being viewed by others
References
Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, Nickerson DA (2003) Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet 33:518–521
Chakraborty R, Jin L (1993) A unified approach to study hypervariable polymorphisms: statistical considerations of determining relatedness and population distances. EXS 67:153–175
Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R (2005) Ascertainment bias in studies of human genome-wide polymorphism. Genome Res 15:1496–1502
Dearlove AM (2002) High throughput genotyping technologies. Brief Funct Genomics Proteomics 1:139–150
Elahi E, Kumm J, Ronaghi M (2004) Global genetic analysis. J Biochem Mol Biol 37:11–27
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14:2611–2620
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred, I: accuracy assessment. Genome Res 8:175–185
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fox S, Filichkin S, Mockler TC (2009) Applications of ultra-high-throughput sequencing. Methods Mol Biol 553:79–108
Harpending H, Rogers A (2000) Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet 1:361–385
LaFramboise T (2009) Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res 37:4181–4193
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Matsumoto T, Okumura N, Uenishi H, Hayashi T, Hamasima N, Awata T (2011) Population structure of pigs determined by single nucleotide polymorphisms observed in assembled expressed sequence tags. Anim Sci J. doi:10.1111/j.1740-0929.2011.00920.x
Mitchell AA, Zwick ME, Chakravarti A, Cutler DJ (2004) Discrepancies in dbSNP confirmation rates and allele frequency distributions from varying genotyping error rates and patterns. Bioinformatics 20:1022–1032
Okumura N, Matsumoto T, Hamasima N, Awata T (2008) Single nucleotide polymorphisms of the KIT and KITLG genes in pigs. Anim Sci J 79:303–313
Okumura N, Hayashi T, Uenishi H, Fukudome N, Komatsuda A, Suzuki A, Shibata M, Nii M, Yamaguchi T, Kojima-Shibata C, Hamasima N, Awata T (2010) Sequence polymorphisms in porcine homologs of murine coat colour-related genes. Anim Genet 41:113–121
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA (2006) Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet 38:375–381
Uenishi H, Eguchi-Ogawa T, Shinkai H, Okumura N, Suzuki K, Toki D, Hamasima N, Awata T (2007) PEDE (Pig EST Data Explorer) has been expanded into Pig Expression Data Explorer, including 10 147 porcine full-length cDNA sequences. Nucleic Acids Res 35:D650–D653
Vicente AA, Carolino MI, Sousa MC, Ginja C, Silva FS, Martinez AM, Vega-Pla JL, Carolino N, Gama LT (2008) Genetic diversity in native and commercial breeds of pigs in Portugal assessed by microsatellites. J Anim Sci 86:2496–2507
Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 34:275–305
Acknowledgments
The authors are greatly indebted to Ikuyo Nakamori (STAFF Institute) for her technical assistance. The authors are also grateful to Taro Obata (STAFF Institute) for administration of the project. This study was supported by the Ministry of Agriculture, Fisheries and Forestry of Japan (Grant number II-304), the Japan Racing Association, and the Livestock Promotion Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Matsumoto, T., Okumura, N., Uenishi, H. et al. Porcine Single Nucleotide Polymorphism (SNP) Development and Population Structure of Pigs Assessed by Validated SNPs. Biochem Genet 50, 428–439 (2012). https://doi.org/10.1007/s10528-011-9486-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-011-9486-9