Abstract
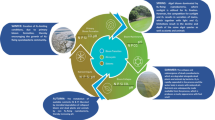
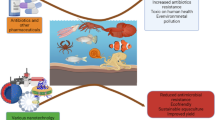
Aquaculture faces significant challenges due to bacterial pathogens like Aeromonas hydrophila, which can severely impact production and fish health. Understanding the relationship between environmental factors, host susceptibility, and bacterial virulence is crucial for effectively managing and mitigating the risks associated with A. hydrophila in aquaculture systems. A. hydrophila, found ubiquitously in aquatic environments, possesses various virulence factors that enhance its pathogenicity. These factors are closely linked to environmental conditions, such as temperature and pH, which play pivotal roles in bacterial growth, survival, and pathogenic potential. Fluctuations in temperature and pH significantly influence A. hydrophila’s metabolic activity and growth rate, thereby modulating its virulence and overall pathogenicity. Ammonia, a byproduct of aquatic organism metabolism and organic matter decomposition, can accumulate to toxic levels in aquaculture settings, compromising fish health and immune function. Elevated ammonia concentrations worsen A. hydrophila infections by compromising host immunity and creating favorable conditions for bacterial proliferation. Oxygen concentration, host signals, and diet formulation are significantly impacting the susceptibility of aquatic organisms to infection. These factors are the most crucial in shaping the ecology, physiology, and pathogenicity of A. hydrophila in aquaculture. There is limited information on how these environmental factors modulate the virulence genes of such important fish pathogens. The knowledge of A. hydrophila virulence and its interplay with environmental factors paves the way for developing strategies to prevent and control diseases in aquaculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Aeromonas belongs to the class of Gamma-proteobacteria and the family Aeromonadaceae (Janda and Abbott 2010). The members of this genus are characterized to be Gram-negative bacilli (0.3–1.0 × 1.0–3.5 µm), positive for oxidase and catalase, and capable of converting nitrates to nitrites, and they are glucose fermenters (Martínez-Murcia et al. 2016). There are several species of aeromonads such as Aeromonas hydrophila that can infect fish and other aquatic animals and spread disease to them (Abdella et al. 2017; Fernández-Bravo and Figueras 2020). Besides being a fish pathogen, it also could cause diseases for humans and can be ubiquitous in aquatic environments, foods, and food processing and storage facilities (Stratev et al. 2012). For instance, diseases caused by Aeromonas spp. in fish are hemorrhagic septicemia, ulcer disease, and motile Aeromonas septicemia (Chen et al. 2019; Zhao et al. 2019). These diseases can result in high mortality rates and high economic losses in aquaculture (Elsheshtawy et al. 2019). Species that infect humans can cause gastroenteritis, wound infections, and septicemia in immunocompromised individuals (Janda and Abbott 2010). In fish, the susceptible organs and tissues include skin, gills, fins, liver, kidney, spleen, and intestine (Wassif and Mohammed 2022). For disease establishment, Aeromonas produce a variety of virulence factors, such as toxins, enzymes, and adhesins, that contribute to their pathogenicity and host damage (Tomás 2012).
The most common species of Aeromonas that cause disease in fish are A. hydrophila, A. sobria, A. veronii, and A. salmonicida (Tomás 2012). The diagnosis of Aeromonas diseases in farmed fish could be based on clinical signs, isolation and identification of the bacteria, molecular methods, and histopathology. Among the aeromonads, A. hydrophila infection continues to be the predominant disease-causing agent in freshwater fish on a global scale (Pattanayak et al. 2020). A. hydrophila was found to have an open pan-genome with a large number of accessory genes and a tiny core genome in a prior comparative genomics investigation (Abdella et al. 2023). The genetic diversity makes it difficult to serotype or genotype the strains of A. hydrophila due to the high complexity in terms of the type and the number of virulence genes present in each isolate (Khor et al. 2015; Abdella et al. 2023). In addition, relying solely on 16 s rRNA gene sequencing for genotyping is impractical in this case(Abdella et al. 2023).
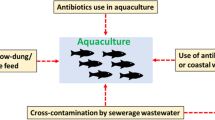
Among different control measures that can applied in the fish farms are antibiotics, vaccines, probiotics, and application of biosecurity measures (Assefa and Abunna 2018). However, the emergence of antibiotic resistance and the diversity of Aeromonas strains pose challenges for the effective control of these diseases (Sherif and Kassab 2023). Resistance to tetracycline antibiotics, such as oxytetracycline and doxycycline, is commonly observed in A. hydrophila isolates from aquaculture context. These antibiotics are extensively being used in fish farming for disease prevention and treatment (Goudarztalejerdi et al. 2022). Additionally, A. hydrophila has shown an increasing resistance trend to fluoroquinolones like ciprofloxacin and enrofloxacin (Nasser et al. 2022). Furthermore, resistance to sulfonamides and trimethoprim, often administered together, has been documented in A. hydrophila strains isolated from aquaculture (Thaotumpitak et al. 2023). Certain A. hydrophila isolates also demonstrate resistance to beta-lactam antibiotics like ampicillin and cephalosporins, widely utilized in human and veterinary medicine (Saengsitthisak et al. 2020; Abdella et al. 2021). Additionally, some A. hydrophila strains exhibit resistance to macrolide antibiotics, including erythromycin and azithromycin (Saengsitthisak et al. 2020). These observations underscore the importance of developing new approaches of effective disease management in aquaculture environments which brings up the importance of the environmental factors.
Environmental factors such as metal availability, salinity, dissolved oxygen concentration, pH value, and temperature as well as poor management (malnutrition, overfeeding, and overcrowding) in hatchery facilities may cause stress to cultured animals and turn it more susceptible to infections by A. hydrophilia (Abreu et al. 2018; Sherif and Kassab 2023). Moreover, A. hydrophila genomes are encoding different types of virulence factors such as aerolysin (aer), cytotoxic enterotoxin (act), heat-stable hemolysin (hly), and heat-stable cytotoxic enterotoxin (ast) (Rasmussen-Ivey et al. 2016; Samayanpaulraj et al. 2020; Sherif and Kassab 2023). These are the most studied virulence determinants in A. hydrophila; however, there are others which might also play a role in its pathogenicity, such as lipase, elastase, flagella, type III secretion system, outer membrane proteins, and DNases (Rasmussen-Ivey et al. 2016; Yassen et al. 2021; Sherif and Kassab 2023). The expression and regulation of these genes may vary depending on the bacterial strain, host, and environmental conditions. It is not fully understood how environmental conditions have an effect on intrinsic bacterial characteristics and the expression of virulence genes, so it is an open question remaining to be elucidated (Abreu et al. 2018; Arayes et al. 2021). Thus, understanding the diversity, function, and environmental regulation of these genes is crucial for developing effective strategies to prevent and treat A. hydrophila infections in fish farms.
Physical factors
Physical factors influence the microbial growth rate, such as temperature and pH value. However, little is known about how these factors can modify the expression of the virulence genes in the pathogenic microbes. It is critical to understand what triggers virulence factor expression or at least the most essential one in a specific pathogen.
Temperature
Temperature is crucial for disease outbreaks in fish. In response to the change of the temperature, bacterial cells need to switch their physiology to tackle the stress and stimuli by the environment (Yi et al. 2022). The optimal temperature for bacterial pathogens in higher animals, or more precisely in endothermic species, will logically be the same as the host body temperature (Cascarano et al. 2021), while in ectothermic hosts, such as fish, the change in temperature of the environment makes the expression of the virulence gene more temperature-dependent and being influenced by the ambient environment temperature. Temperature-dependent expression of virulence genes in Aeromonas in fish has been reported for some genes, such as aerolysin (aer), cytotoxic enterotoxin (act), hemolysin A (hylA), outer membrane protein (omp), elastase, flagellin, lipase, and type III secretion system (T3SS) (Pattanayak et al. 2020). However, the molecular mechanisms and regulatory pathways involved in this process are not fully understood. Adding to that, the increase in temperature is found to be stressful to the host, which increases the expression of corticosteroids which increase the vulnerability of the fish for infection (Huizinga et al. 1979).
In Aeromonas salmonicida SRW-OG1, a mesophilic fish pathogen, type 2 secretion system (T2SS) has a role in secreting degradative enzymes and contributes directly to the pathogenesis. The expression virulence factors such as protease, lipase, and lecithinase enzyme as well as the hemolytic ability were modulated by the temperature changes (Yi et al. 2022). The activity of protease was the highest at 18 °C compared to 37 °C, while the lipase activity was the highest at elevated temperature at 28 and 37 °C in comparison to lower expression detected at 18 °C (Yi et al. 2022). Lecithinase activity was the lowest at 37 °C (Yi et al. 2022). In transcriptome analysis of Aeromonas salmonicida, different temperatures representing 3 different temperatures, namely, 18 °C, 28 °C, and 37 °C, defined the optimum temperature for cold-water fish pathogen, warm-water fish, and humans’ pathogens, respectively. The results revealed that aerA and hlyA genes were found to be significantly upregulated at 28 and 37 °C.
Another study conducted by Pattanayak et al. (2020) on the carp Labeo rohita assessed the influence of different temperature on virulence gene expression in A. hydrophila in vivo. The result showed that the studied gene expression was highest at 28 °C, rather than 37 °C. The highest virulence gene expressed in A. hydrophila at 28 °C was aerolysin, cytotoxic enterotoxin, elastase, hemolysin, flagellin, outer membrane protein, and T3SS. However, this finding cannot be generalized as the number of virulence genes is not fixed among the A. hydrophila strains besides that the study was limited to the carp Labeo rohita as a model in which the result might vary with host variation. In other words, the expression of virulence genes in Aeromonas can vary depending on the water temperature and the host animal (Pattanayak et al. 2020; Sadique et al. 2021). The inconsistency of the expression patterns of many virulence factors in the same strain in response to temperature changes emphasizes the difficulty and the need of identifying the major virulence factor that is responsible for disease development in the susceptible host.
Ammonia and pH
Ammonia and pH are closely connected in fish farming. Ammonia is a nitrogenous waste product originated from fish metabolism, and its concentration and its ionization state in water are governed by the pH levels and temperature (Yavuzcan Yildiz et al. 2017). The recommended pH range for tilapia fish farming ranged from 6.5 to 9.5, with the optimum pH level falling between 6.5 and 8.5. pH levels outside this range, either higher or lower, can cause stress to the fish (Boyd 2020; Francis-Floyd et al. 2022). At optimum pH levels, most of the ammonia is present in the form of ammonium ions, which are less toxic to fish. However, a rise in the pH levels above 10 increases the concentration of unionized ammonia, posing substantial risks to fish by negatively impacting their productivity, health, and potentially leading to mortality (Bhatnagar and Devi 2013; Hasim et al., 2017).
Changes in pH and ammonia concentration may have an impact not only on the welfare of the fish but also on the expression of virulence genes by pathogens present in the system. The impacts of pH and ammonia on the activation of Aeromonas virulence genes were observed. Abreu et al. (2018) reported that high mortality rates were observed in tilapia challenged with A. hydrophila within an environment characterized by high pH levels and ammonia concentrations for each parameter when tested individually. Furthermore, their study in vitro revealed a notable upregulation in the expression of the fla gene when A. hydrophila was cultured in an environment with elevated ammonia concentration compared to other virulence genes (aerolysin and lipase) while pH-tested groups showed no expression of any virulence genes (Abreu et al. 2018). This study declares the potential role of ammonia as a crucial factor for A. hydrophila virulence, which may contribute to the proliferation of this pathogenic bacterium. Moreover, it was known that an increase in pH levels may act to suppress the immune system of the fish, rendering them more susceptible to opportunistic pathogens. Nevertheless, it is worth emphasizing that while these findings shed light on the general relationship between environmental factors, virulence gene expression in A. hydrophila, and their impact on fish, the study did not provide a detailed elucidation of gene expression patterns. In recent study conducted by Abdel-Latif et al. (2022), it was found that the presence of total ammonia-N and A. hydrophila together exaggerated their effects on Nile tilapia to produce a severe disease followed by high mortality rates. Therefore, this comprehensive study provides valuable mechanistic insights into the ramifications of fish exposure to ammonia stress and infection with A. hydrophila, shedding light on the intricacies of this interaction. Also, it was found that virulence genes of A. hydrophila specifically aerA and hly genes were expressed under conditions of water quality characterized by elevated levels of unionized ammonia with infection by the parasite Gyrodactylus cichlidarum in Nile tilapia (Abdel-Latif and Khafaga 2020). In-depth studies are warranted to elucidate the intricate interplay between the expression of virulence genes in A. hydrophila and the specific pH and ammonia parameters, with direct implications for fish health and susceptibility.
Oxygen concentration
Aeromonads are facultative anaerobes, and as all other facultative microbes, they may switch between aerobic and anaerobic metabolism. Such switching behaviors are controlled by gene expression and are triggered by molecular sensing of oxygen concentration. No surprise, it has developed a molecular mechanism for doing so through gene expression control, including virulence genes. Oxygen can regulate virulence genes by modulating the activity of transcription factors—which are proteins that bind to specific DNA sequences and control gene expression. One example of a transcription factor that responds to oxygen is fumarate and nitrate reductase regulator (FNR), which is activated under low oxygen conditions and represses genes involved in aerobic respiration and activates genes involved in anaerobic respiration and virulence (Barbieri et al. 2014). The active form of FNR is homodimer protein and requires an iron-sulfur cluster in the FNR [4Fe-4S]2+ configuration. The cluster transformed into [2Fe-2S]2+ form in the presence of oxygen, resulting in dimer dissociation and FNR inactivation; the process of FNR activation and deactivation is reviewed by Barbieri et al. (2014). Furthermore, FNR activation is very crucial for human pathogens such as Salmonella enterica, Pseudomonas aeruginosa, Neisseria meningitidis, and Escherichia coli for survival in low oxygen environments (Contreras et al. 1997; Fink et al. 2007; Kushwaha et al. 2020).
Another example is nitrate/nitrite response regulator (NarL), which is activated by nitrate or nitrite and regulates genes involved in nitrate reduction and virulence (Goh et al. 2005; Durand and Guillier 2021). Oxygen can also affect the expression of virulence genes indirectly by influencing the production of reactive oxygen species (ROS) and nitric oxide (NO), which are molecules that can damage bacterial cells and are part of the host immune response (Fajardo et al. 2023). Some bacteria have mechanisms to sense and detoxify ROS and NO, such as catalases, superoxide dismutase, and NO reductases. These enzymes can also modulate the expression of virulence genes by affecting the redox state of the cell or by interacting with transcription factors. Therefore, oxygen may be considered as a key environmental factor that influences the expression of virulence genes in many bacterial strains including A. hydrophila and their ability to cause disease. However, there has been little research into the effect of oxygen concentrations on the expression of virulence genes in fish infections. Figure 1 depicts the interplay between the environment, microbes, and host.
Host signals
The effect of host signals on the expression of A. hydrophila virulence genes and how it is modulated by such signals cannot be understood without challenging the pathogens to host tissue. For example, a proteomics investigation on the differential expression of pathogenic E. coli cultivated on blood agar and solid medium indicated that cell growth on blood agar has increased genes important for anaerobic fermentation pathways as well as several virulence features (Pettersen et al. 2016; Vlaeminck et al. 2022). From the previous discussion, it can be concluded that the virulence genes in Aeromonas are also regulated by the presence of host cell components present in blood, serum, and tissue. For instance, cytotoxic toxins such as aerolysin and hemolysin are produced in an inactive form inside the bacterial cell; once activated, they are released to the environment to attack the host cell (Tomás 2012). These components could bind to receptors on the surface of Aeromonas cells and induce the expression of virulence genes (Pettersen et al. 2016). Deeper investigation is necessary to fully understand the mechanisms of activation and interaction between host signals and the regulation of virulence genes in A. hydrophila. The host–pathogen interaction in fish is extremely challenging to research due to the combination and diversity of infection situations and settings. For example, Aeromonas spp. can cause disease in a wide range of aquatic animals in addition to humans, and genetic diversity of Aeromonas spp. causes various strains to have varied growth requirements and optimal environments (Froquet et al. 2007). While initial research models offered a simplified approach for studying host microbe interaction, transcriptomics and proteomics studies enable investigation of the real host–pathogen interaction, yielding more reliable results. However, this approach can be cost-intensive, time-consuming, and technically demanding. Additionally, the findings are specific to the studied host and pathogen, limiting generalizability.
Organic matter and virulence
The formulation of fish diets has a considerable impact on microbial ecology in the aquatic environment, as well as microbial pathogenicity, interaction, and abundance (Qin et al. 2016; Zhang et al. 2020). A. hydrophila infection is associated with water with a high organic load, and nutrient-rich feed pollutes aquatic water systems (Semwal et al. 2023). Motile Aeromonas septicemia outbreaks in crucian carp in China were also shown to have occurred in ponds with high organic loads, indicating that excessive nutrient loading owing to fertilizers and algal blooming into aquatic environments is a major source of eutrophication (Lu et al. 2019). As reported by Zhang et al. (2020), in ponds where excessive feed was provided, the prevalence of Aeromonas spp. was notably high. This was particularly concerning because when excess unconsumed feeds were left in the water under specific conditions, it could lead to the rapid multiplication of virulent A. hydrophila over a short period of time. If the density of A. hydrophila reached a critical level, such as 108 CFU/mL, a significant incidence of fish mortality was observed. Although the particular nutrient that made the bacteria thrive in fish farming ponds and produce disease outbreaks is unknown, it is also uncertain which nutrients specifically contribute to A. hydrophila’s virulence (Abreu et al. 2018; Zhang et al. 2020). Without ignoring the fact that aeromonads prefer ammonium as a source of nitrogen, which is produced by organic wastes remaining in the bonds, it appears today that the unutilized diet contributes to an increase in the number of aeromonads in the pond water.
The precise formulation of the fish feed and its utilization by fish is very critical in controlling the infection and suppressing the activation of virulence genes in A. hydrophila. For instance, NSS microalgae mixture containing Nannochloropsis oculate, Schizochytrium, and Spirulina species was effective against the infection with A. hydrophila. Remarkably, higher levels of NSS microalgae blend in Nile tilapia feed supplements exhibited lower mortality rates that came in harmony with the positive reduction in A. hydrophila counts (Ibrahim et al. 2022). These improved survival rates could be assigned to the favorable effects of NSS microalgae mixture on both antioxidant and immune activities of Nile tilapia, therefore recommending that feeding of microalgae enhances the fish immune response (Najdenski et al. 2013).
More research is needed to definitely confirm the link between various dietary components and virulence gene overexpression in A. hydrophila. However, feeding fish a diet high in omega-3 fatty acids can assist to improve the immune system and make the fish less susceptible to infection; this relationship is not fully understood.
Conclusion
In conclusion, the environmental regulation of virulence in A. hydrophila emerges as a complex and dynamic interplay between the bacterium and its environment. Temperature, dissolved oxygen, pH, ammonia, and nutrient availability act as a modulator, coordinating the expression of virulence genes and toxin production. This remarkable adaptability allows A. hydrophila to thrive in diverse aquatic environments and establish infections across a range of hosts, posing a significant challenge to aquaculture. Unraveling the mechanisms of environmental virulence regulation presents a compelling target for future research. By elucidating these pathways, we can pave the way for the development of targeted interventions to mitigate A. hydrophila infections, ultimately promoting a healthier and more sustainable aquaculture industry.
Data availability
No datasets were generated or analyzed during the current study.
References
Abdella B, El-Wazzan E, El-Sersy NA et al (2017) Pathogenicity and antibiotic susceptibility of two bacterial pathogens associated with the clam Tapes decussatus in some Egyptian fisheries. Ege J Fish Aquat Sci 34:383–389. https://doi.org/10.12714/egejfas.2017.34.4.04
Abdella B, Abozahra NA, Shokrak NM et al (2023) Whole spectrum of Aeromonas hydrophila virulence determinants and the identification of novel SNPs using comparative pathogenomics. Sci Rep 13:7712. https://doi.org/10.1038/s41598-023-34887-1
Abdella M, Abdella B, Lahiri C (2021) Rediscovering and repurposing natural microbial macromolecules through computational approaches. In: Das S, Dash HRBT-M and NM (eds) Microbial and Natural Macromolecules. Elsevier, pp 373–400
Abdel-Latif HMR, Khafaga AF (2020) Natural co-infection of cultured Nile tilapia Oreochromis niloticus with Aeromonas hydrophila and Gyrodactylus cichlidarum experiencing high mortality during summer. Aquac Res 51:1880–1892. https://doi.org/10.1111/are.14538
Abdel-Latif HMR, Shukry M, Abd-elaziz RA (2022) Clinico-pathological findings and expression of inflammatory cytokines, apoptosis, and oxidative stress-related genes draw mechanistic insights in Nile tilapia reared under ammonia-N exposure and Aeromonas hydrophila challenge. Fish Shellfish Immunol 127:1–12. https://doi.org/10.1016/j.fsi.2022.06.001
Abreu REFF, Magalhães TC, Souza RC et al (2018) Environmental factors on virulence of Aeromonas hydrophila. Aquac Int 26:495–507. https://doi.org/10.1007/s10499-017-0230-2
Arayes MA, Mabrouk MEM, Sabry SA, Abdella B (2021) Diversity and characterization of culturable haloalkaliphilic bacteria from two distinct hypersaline lakes in northern Egypt. Biologia (bratisl) 76:751–761. https://doi.org/10.2478/s11756-020-00609-5
Assefa A, Abunna F (2018) Maintenance of fish health in aquaculture: review of epidemiological approaches for prevention and control of infectious disease of fish. Vet Med Int 2018:1–10. https://doi.org/10.1155/2018/5432497
Barbieri NL, Nicholson B, Hussein A et al (2014) FNR regulates expression of important virulence factors contributing to pathogenicity of uropathogenic Escherichia coli. Infect Immun 82:5086–5098. https://doi.org/10.1128/IAI.02315-14
Bhatnagar A, Devi P (2013) Water quality guidelines for the management of pond fish culture. Int J Environ Sci 3:1980–2009. https://doi.org/10.6088/ijes.2013030600019
Boyd CE (2020). Water Quality. In Springer Nature Switzerland AG (Third, Vol. 3, Issue Rosborg 2015). Springer International Publishing. https://doi.org/10.1007/978-3-030-23335-8
Cascarano MC, Stavrakidis-Zachou O, Mladineo I, et al (2021) Mediterranean aquaculture in a changing climate: temperature effects on pathogens and diseases of three farmed fish species. Pathogens 10 https://doi.org/10.3390/pathogens10091205
Chen F, Sun J, Han Z, et al (2019) Isolation, identification and characteristics of Aeromonas veronii from diseased crucian carp (Carassius auratus gibelio). Front Microbiol 10 https://doi.org/10.3389/fmicb.2019.02742
Contreras I, Toro CS, Troncoso G, Mora GC (1997) Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143:2665–2672. https://doi.org/10.1099/00221287-143-8-2665
Durand S, Guillier M (2021) Transcriptional and post-transcriptional control of the nitrate respiration in bacteria. Front Mol Biosci 8 https://doi.org/10.3389/fmolb.2021.667758
Elsheshtawy A, Yehia N, Elkemary M, Soliman H (2019) Investigation of Nile tilapia summer mortality in Kafr El-Sheikh governorate. Egypt Genet Aquat Org 3:17–25. https://doi.org/10.4194/2459-1831-v3_1_03
Fajardo C, Santos P, Passos R et al (2023) Early molecular immune responses of turbot (Scophthalmus maximus L.) following infection with Aeromonas salmonicida subsp. salmonicida. Int J Mol Sci 24:1–15. https://doi.org/10.3390/ijms241612944
Fernández-Bravo A, Figueras MJ (2020) An update on the genus aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms 8(1):129. https://doi.org/10.3390/microorganisms8010129
Fink RC, Evans MR, Porwollik S et al (2007) FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol 189:2262–2273. https://doi.org/10.1128/JB.00726-06
Francis-Floyd R, Watson C, Petty D, Pouder D (2022) Ammonia in aquatic systems. EDIS 2022:1–6. https://doi.org/10.32473/edis-fa031-2022
Froquet R, Cherix N, Burr SE et al (2007) Alternative host model to evaluate Aeromonas virulence. Appl Environ Microbiol 73:5657–5659. https://doi.org/10.1128/AEM.00908-07
Goh E-B, Bledsoe PJ, Chen L-L et al (2005) Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J Bacteriol 187:4890–4899. https://doi.org/10.1128/JB.187.14.4890-4899.2005
Goudarztalejerdi A, Yavari M, Kalourazi MN, et al (2022) Antibiotic resistance and virulence factor gene profile of A. hydrophila isolated from carp (Cyprinidae) suspected with hemorrhagic septicemia in Gilan, Iran. https://doi.org/10.1111/lam.13806
Hasim KY, Kasim F (2017) Suitable location map of floating net cage for environmentally friendly fish farming development with geographic information systems applications in lake Limboto, Gorontalo, Indonesia. AACL Bioflux 10:254–264
Huizinga HW, Esch GW, Hazen TC (1979) Histopathology of red-sore disease (Aeromonas hydrophila) in naturally and experimentally infected largemouth bass Micropterus salmoides (Lacepede). J Fish Dis 2:263–277. https://doi.org/10.1111/j.1365-2761.1979.tb00169.x
Ibrahim D, Abd El-Hamid MI, Al-Zaban MI et al (2022) Impacts of fortifying Nile tilapia (Oreochromis niloticus) diet with different strains of microalgae on its performance, fillet quality and disease resistance to Aeromonas hydrophila considering the interplay between antioxidant and inflammatory response. Antioxidants 11:2181. https://doi.org/10.3390/antiox11112181
Janda JM, Abbott SL (2010) The genus Aeromonas : taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73. https://doi.org/10.1128/CMR.00039-09
Khor WC, Puah SM, Tan JAMA et al (2015) Phenotypic and genetic diversity of Aeromonas species isolated from fresh water lakes in Malaysia. PLoS ONE 10:1–13. https://doi.org/10.1371/journal.pone.0145933
Kushwaha SK, Bhavesh NLS, Abdella B et al (2020) The phylogenomics of CRISPR-Cas system and revelation of its features in Salmonella. Sci Rep 10:1–13. https://doi.org/10.1038/s41598-020-77890-6
Lu J, Zhu B, Struewing I et al (2019) Nitrogen–phosphorus-associated metabolic activities during the development of a cyanobacterial bloom revealed by metatranscriptomics. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-38481-2
Martínez-Murcia A, Beaz-Hidalgo R, Navarro A et al (2016) Aeromonas lusitana sp. nov., isolated from untreated water and vegetables. Curr Microbiol 72:795–803. https://doi.org/10.1007/s00284-016-0997-9
Najdenski HM, Gigova LG, Iliev II et al (2013) Antibacterial and antifungal activities of selected microalgae and cyanobacteria. Int J Food Sci Technol 48:1533–1540. https://doi.org/10.1111/ijfs.12122
Nasser H, Abdeltawab A, Maroof A, Elkhyat M (2022) Molecular characterization and antimicrobial effect of some antibiotics on Aeromonas hydrophila isolated from different sources. Benha Vet Med J 43:45–50. https://doi.org/10.21608/bvmj.2022.153716.1567
Pattanayak S, Priyadarsini S, Paul A et al (2020) Diversity of virulence-associated genes in pathogenic Aeromonas hydrophila isolates and their in vivo modulation at varied water temperatures. Microb Pathog 147:104424. https://doi.org/10.1016/j.micpath.2020.104424
Pettersen VK, Mosevoll KA, Lindemann PC, Wiker HG (2016) Coordination of metabolism and virulence factors expression of extraintestinal pathogenic Escherichia coli purified from blood cultures of patients with sepsis. Mol Cell Proteomics 15:2890–2907. https://doi.org/10.1074/mcp.M116.060582
Qin Y, Hou J, Deng M et al (2016) Bacterial abundance and diversity in pond water supplied with different feeds. Sci Rep 6:1–13. https://doi.org/10.1038/srep35232
Rasmussen-Ivey CR, Figueras MJ, McGarey D, Liles MR (2016) Virulence factors of Aeromonas hydrophila: in the wake of reclassification. Front Microbiol 7:1–10. https://doi.org/10.3389/fmicb.2016.01337
Sadique A, Neogi SB, Bashar T, et al (2021) Dynamics, diversity, and virulence of Aeromonas spp. in homestead pond water in coastal Bangladesh. Front Public Heal 9 https://doi.org/10.3389/fpubh.2021.692166
Saengsitthisak B, Chaisri W, Punyapornwithaya V et al (2020) Occurrence and antimicrobial susceptibility profiles of multidrug-resistant aeromonads isolated from freshwater ornamental fish in chiang mai province. Pathogens 9:1–13. https://doi.org/10.3390/pathogens9110973
Samayanpaulraj V, Sivaramapillai M, Palani SN et al (2020) Identification and characterization of virulent Aeromonas hydrophila Ah17 from infected Channa striata in river Cauvery and in vitro evaluation of shrimp chitosan. Food Sci Nutr 8:1272–1283. https://doi.org/10.1002/fsn3.1416
Semwal A, Kumar A, Kumar N (2023) A review on pathogenicity of Aeromonas hydrophila and their mitigation through medicinal herbs in aquaculture. Heliyon 9:e14088. https://doi.org/10.1016/j.heliyon.2023.e14088
Sherif AH, Kassab AS (2023) Multidrug-resistant Aeromonas bacteria prevalence in Nile tilapia broodstock. BMC Microbiol 23:80. https://doi.org/10.1186/s12866-023-02827-8
Stratev D, Vashin I, Rusev V (2012) Characteristics of bacteria from the genus Aeromonas prevalence and survival of Aeromonas spp. in foods-a review. Rev Méd Vét 163:486–494
Thaotumpitak V, Sripradite J, Atwill ER, Jeamsripong S (2023) Emergence of colistin resistance and characterization of antimicrobial resistance and virulence factors of Aeromonas hydrophila, Salmonella spp., and Vibrio cholerae isolated from hybrid red tilapia cage culture. PeerJ 11 https://doi.org/10.7717/peerj.14896
Tomás JM (2012) The main Aeromonas pathogenic factors. ISRN Microbiol 2012:1–22. https://doi.org/10.5402/2012/256261
Vlaeminck J, Lin Q, Xavier BB et al (2022) The dynamic transcriptome during maturation of biofilms formed by methicillin-resistant Staphylococcus aureus. Front Microbiol 13:1–15. https://doi.org/10.3389/fmicb.2022.882346
Wassif I, Mohammed R (2022) Use of thyme and thymol as immunostimulant agents to control experimental Aeromonas hydrophyla infection in Nile tilapia (Oreochromis niloticus). Zagazig Vet J 50:241–254. https://doi.org/10.21608/zvjz.2022.150326.1185
Yassen M, EL-Naenaeey E-S, Omar A, Tartor Y (2021) Virulence determinants of Aeromonas species implicated in fish diseases and control of infection: an overview. Zagazig Vet J 49:284–299. https://doi.org/10.21608/zvjz.2021.85132.1148
Yavuzcan Yildiz H, Robaina L, Pirhonen J et al (2017) Fish welfare in aquaponic systems: its relation to water quality with an emphasis on feed and faeces—a review. Water 9:13. https://doi.org/10.3390/w9010013
Yi X, Chen Y, Cai H et al (2022) The temperature-dependent expression of type II secretion system controls extracellular product secretion and virulence in mesophilic Aeromonas salmonida SRW-OG1. Front Cell Infect Microbiol 12:1–16. https://doi.org/10.3389/fcimb.2022.945000
Zhang D, Xu DH, Shoemaker CA, Beck BH (2020) The severity of motile Aeromonas septicemia caused by virulent Aeromonas hydrophila in channel catfish is influenced by nutrients and microbes in water. Aquaculture 519:734898. https://doi.org/10.1016/j.aquaculture.2019.734898
Zhao X-L, Jin Z-H, Di G-L et al (2019) Molecular characteristics, pathogenicity and medication regimen of Aeromonas hydrophila isolated from common carp (Cyprinus carpio L.). J Vet Med Sci 81:1769–1775. https://doi.org/10.1292/jvms.19-0025
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Yomna M. Elshamy was responsible for data collection and analysis and the writing of the first draft. Hossam I. Kadira was responsible for data collection and analysis and the writing of the first draft. Nermeen M. Shokrak was responsible for the conceptualization and editing after reviews. Nourhan A. Abozahra was responsible for editing and proofreading. Radi A. Mohamed was responsible for editing and proofreading. Bahaa Abdella conceptualized, edited, reviewed, and finalized the manuscript. All the authors read and approved the final draft of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies on humans or on animals.
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Amany Abbass
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdella, B., Shokrak, N.M., Abozahra, N.A. et al. Aquaculture and Aeromonas hydrophila: a complex interplay of environmental factors and virulence. Aquacult Int (2024). https://doi.org/10.1007/s10499-024-01535-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10499-024-01535-y