Abstract
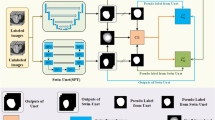
Medical image segmentation is a crucial task in many clinical applications, such as tumor detection and surgical planning. However, the annotation process for medical images is often both time-consuming and expensive, which requires professional knowledge and experience. This study aims to develop a medical image segmentation method based on semi-supervised learning, which can improve the performance of segmentation by effectively using labeled data and unlabeled data. We proposed a novel multi-task semi-supervised method based on multi-branch cross pseudo supervision, called MS2MPS, which can efficiently utilize unlabeled data for semi-supervised medical image segmentation. The proposed method consists of two multi-task backbone networks with multiple output branches, which were used to simultaneously generate segmentation probability maps (SPM) and signed distance maps (SDM) to get more constraints and information. Moreover, a multi-branch cross pseudo supervised (MPS) approach was proposed to promote the high similarity between predictions of the same input image by multiple perturbation networks. Experiments on the public medical image segmentation dataset Automated Cardiac Diagnosis Challenge (ACDC) and Left Atrium (LA) dataset demonstrate that our approach is superior to existing some semi-supervised segmentation methods in terms of Dice similarity coefficient (DSC), 95% Hausdorff distance (HD95), and Jaccard Similarity Coefficient (JSC). When trained with 10% labeled data, compared to the best results in all compared methods, our approach improved the DSC by 2.63% and 1.54% on the ACDC and LA datasets, increased the JSC by 23.15% and 0.79%, and simultaneously reduced the HD95 by 12.57% and 7.77% on the respective datasets. Our proposed semi-supervised medical image segmentation approach is an effective and practical solution for medical image analysis, particularly in scenarios where labeled data is limited or expensive.








Similar content being viewed by others
References
Li J, Udupa JK, Tong Y, Wang L, Torigian DA (2020) LinSEM: linearizing segmentation evaluation metrics for medical images. Med Image Anal 60:101601. https://doi.org/10.1016/j.media.2019.101601
Kim BN, Dolz J, Jodoin PM, Desrosiers C (2021) Privacy-Net: an adversarial approach for Identity-Obfuscated segmentation of medical images. IEEE Trans Med Imaging 40(7):1737–1749. https://doi.org/10.1109/TMI.2021.3065727
Iqbal A, Sharif M, Yasmin M, Raza M, Aftab S (2022) Generative adversarial networks and its applications in the biomedical image segmentation: a comprehensive survey. Int J Multimed Inf Retr 11(3):333–368. https://doi.org/10.1007/s13735-022-00240-x
Krishnan R, Rajpurkar P, Topol EJ (2022) Self-supervised learning in medicine and healthcare. Nat Biomed Eng 6(12):1346–1352. https://doi.org/10.1038/s41551-022-00914-1
Cheplygina V, de Bruijne M, Pluim J (2019) Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal 54:280–296. https://doi.org/10.1016/j.media.2019.03.009
Noroozi M, Favaro P (2016) Unsupervised learning of visual representations by solving jigsaw puzzles. In: Computer vision - ECCV 2016, Springer, 2016, pp 69-84
Berthelot D, Carlini N, Goodfellow I, Papernot N, Oliver A, Raffel CA (2019) Mixmatch: A holistic approach to semi-supervised learning. Adv Neural Inf Process Syst 32. https://doi.org/10.48550/arXiv.1905.02249
Chaitanya K, Erdil E, Karani N, Konukoglu E (2023) Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation. Med Image Anal 102792. https://doi.org/10.1016/j.media.2023.102792
Wang Y, Wang H, Shen Y, Fei J, Li W, Jin G, Wu L, Zhao R, Le X (2022) Semi-supervised semantic segmentation using unreliable pseudo-labels. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition 2022:4248–4257
Wang Q, Li X, Chen M, Chen L, Chen J (2022) A regularization-driven Mean Teacher model based on semi-supervised learning for medical image segmentation. Phys Med Biol 67(17):175010
Wu L, Li J, Wang Y, Meng Q, Qin T, Chen W, Zhang M, Liu T (2021) R-drop: regularized dropout for neural networks. Adv Neural Inf Process Syst 34:10890–10905
Li J, Speier W, Ho KC, Sarma KV, Gertych A, Knudsen BS, Arnold CW (2018) An EM-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies. Comput Med Imag Grap 69:125–133
Zheng H, Lin L, Hu H, Zhang Q, Chen Q, Iwamoto Y, Han X, Chen Y, Tong R, Wu J (2019) Semi-supervised segmentation of liver using adversarial learning with deep atlas prior. Medical image computing and computer assisted intervention-MICCAI 2019: 22nd international conference. Springer, Shenzhen, China, pp 148–156
Yao H, Hu X, Li X (2022) Enhancing pseudo label quality for semi-supervised domain-generalized medical image segmentation. In: Proceedings of the AAAI conference on artificial intelligence, pp 3099–3107
Bortsova G, Dubost F, Hogeweg L, Katramados I, De Bruijne M (2019) Semi-supervised medical image segmentation via learning consistency under transformations. Medical image computing and computer assisted intervention-MICCAI 2019: 22nd international conference. Springer, Shenzhen, China, pp 810–818
Hu X, Zeng D, Xu X, Shi Y (2021) Semi-supervised contrastive learning for label-efficient medical image segmentation. Medical image computing and computer assisted intervention-MICCAI 2021: 24th international conference. Springer, Strasbourg, France, pp 481–490
Liu X, Hu Y, Chen J, Li K (2022) Shape and boundary-aware multi-branch model for semi-supervised medical image segmentation. Comput Biol Med 143: 105252. https://doi.org/10.1016/j.compbiomed.2022.105252
Karimi D, Salcudean SE (2019) Reducing the hausdorff distance in medical image segmentation with convolutional neural networks. IEEE T Med Imaging 39(2):499–513. https://doi.org/10.1109/TMI.2019.2930068
Tamal M (2020) Intensity threshold based solid tumour segmentation method for Positron Emission Tomography (PET) images: a review. Heliyon 6(10):e5267. https://doi.org/10.1016/j.heliyon.2020.e05267
Javadpour A, Mohammadi A (2016) Improving brain magnetic resonance image (MRI) segmentation via a novel algorithm based on genetic and regional growth. J Biomed Phys Eng 6(2):95–108
Jordan MI, Mitchell TM (2015) Machine learning: trends, perspectives, and prospects. Science 349(6245):255–260
Wang S, Yang DM, Rong R, Zhan X, Xiao G (2019) Pathology image analysis using segmentation deep learning algorithms. Am J Pathol 189(9):1686–1698. https://doi.org/10.1016/j.ajpath.2019.05.007
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Medical image computing and computer-assisted intervention-MICCAI 2015: 18th international conference. Springer, Munich, Germany, pp 234–241
Abdollahi A, Pradhan B, Alamri A (2020) VNet: an end-to-end fully convolutional neural network for road extraction from high-resolution remote sensing data. IEEE Access 8:179424–179436. https://doi.org/10.1109/ACCESS.2020.3026658
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2019) Unet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE T Med Imaging 39(6):1856–1867. https://doi.org/10.1109/TMI.2019.2959609
çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. Medical image computing and computer-assisted intervention-MICCAI 2016: 19th international conference. Springer, Athens, Greece, pp 424–432
Fan L, Zhao H, Li Y, Li S, Zhou R, Chu W (2022) RAO-UNet: a residual attention and octave UNet for road crack detection via balance loss. IET Intell Transp Sy 16(3): 332–343. https://doi.org/10.1049/itr2.12146
Li W, Qin S, Li F, Wang L (2021) MAD-UNet: a deep U-shaped network combined with an attention mechanism for pancreas segmentation in CT images. Med Phys 48(1):329–341. https://doi.org/10.1002/mp.14617
Zhou Y, Huang W, Dong P, Xia Y, Wang S (2019) D-UNet: a dimension-fusion U shape network for chronic stroke lesion segmentation. IEEE/ACM Trans Comput Biol Bioinforma 18(3):940–950. https://doi.org/10.1109/TCBB.2019.2939522
Li S, Liu N, Li F, Gao J, Ding J (2022) Automatic fault delineation in 3-D seismic images with deep learning: data augmentation or ensemble learning? IEEE T Geosci Remote 60:1–14. https://doi.org/10.1109/TGRS.2022.3150353
Christoffersen P, Jacobs K (2004) The importance of the loss function in option valuation. J Financ Econ 72(2): 291–318. https://doi.org/10.1016/j.jfineco.2003.02.001
Köksoy O (2006) Multiresponse robust design: Mean square error (MSE) criterion, Appl. Math. Comput. 175(2):1716–1729. https://doi.org/10.1016/j.amc.2005.09.016
Huttenlocher DP, Klanderman GA, Rucklidge WJ (1993) Comparing images using the Hausdorff distance. IEEE T Pattern Anal 15(9):850–863. https://doi.org/10.1109/34.232073
Tougaard S, Chorkendorff I (1987) Differential inelastic electron scattering cross sections from experimental reflection electron-energy-loss spectra: Application to background removal in electron spectroscopy. Phys Rev B 35(13): 6570. https://doi.org/10.1103/PhysRevB.35.6570
Ho Y, Wookey S (2019) The real-world-weight cross-entropy loss function: modeling the costs of mislabeling. IEEE access 8:4806–4813. https://doi.org/10.1109/ACCESS.2019.2962617
Lin T, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, pp 2980–2988
Tarvainen A, Valpola H (2017) Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results, Advances in neural information processing systems 30: 1672. https://ieeexplore.ieee.org/book/6267330
Bortsova G, Dubost F, Hogeweg L, Katramados I, De Bruijne M (2019) Semi-supervised medical image segmentation via learning consistency under transformations. Medical image computing and computer assisted intervention-MICCAI 2019: 22nd international conference. Springer, Shenzhen, China, pp 810–818
Mittal S, Tatarchenko M, Brox T (2019) Semi-supervised semantic segmentation with high-and low-level consistency. IEEE T Pattern Anal 43(4):1369–1379. https://doi.org/10.1109/TPAMI.2019.2960224
Luo X, Liao W, Chen J, Song T, Chen Y, Zhang S, Chen N, Wang G, Zhang S (2021) Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency. In: Medical image computing and computer assisted intervention-MICCAI 2021: 24th international conference, Springer, pp 318–329
Grandvalet Y, Bengio Y (2004) Semi-supervised learning by entropy minimization. Adv Neural Inf Process Syst 17: 529–536. https://doi.org/10.5555/2976040.2976107
Miyato T, Maeda S, Koyama M, Ishii S (2018) Virtual adversarial training: A regularization method for supervised and semi-supervised learning. IEEE T Pattern Anal 41(8):1979–1993. https://doi.org/10.1109/TPAMI.2018.2858821
Fan J, Gao B, Jin H, Jiang L (2022) Ucc: uncertainty guided cross-head co-training for semi-supervised semantic segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 9947-9956
Wang K, Zhan B, Zu C, Wu X, Zhou J, Zhou L, Wang Y (2022) Semi-supervised medical image segmentation via a tripled-uncertainty guided mean teacher model with contrastive learning. Med Image Anal 79: 102447. https://doi.org/10.1016/j.media.2022.102447
Lei T, Zhang D, Du X, Wang X, Wan Y, Nandi AK (2022) Semi-supervised medical image segmentation using adversarial consistency learning and dynamic convolution network. IEEE Trans Med Imaging PP. https://doi.org/10.1109/TMI.2022.3225687
Luo X, Wang G, Liao W, Chen J, Song T, Chen Y, Zhang S, Metaxas DN, Zhang S (2022) Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med Image Anal 80: 102517. https://doi.org/10.1016/j.media.2022.102517
Tan Z, Li S, Hu Y, Tao H, Zhang L (2023) Semi-XctNet: volumetric images reconstruction network from a single projection image via semi-supervised learning. Comput Biol Med 155: 106663. https://doi.org/10.1016/j.compbiomed.2023.106663
Wu W, Yan J, Zhao Y, Sun Q, Zhang H, Cheng J, D. Liang, Y. Chen, Z. Zhang, Z. Li, Multi-task learning for concurrent survival prediction and semi-supervised segmentation of gliomas in brain MRI, Displays 78 (2023) 102402, https://doi.org/https://doi.org/10.1016/j.displa.2023.102402
Chen X, Yuan Y, Zeng G, Wang J (2021) Semi-supervised semantic segmentation with cross pseudo supervision. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 2613–2622
Ouali Y, Hudelot C, Tami M (2020) Semi-supervised semantic segmentation with cross-consistency training. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 12674–12684
Li S, Zhang C, He X (2020) Shape-aware semi-supervised 3D semantic segmentation for medical images. In: Medical image computing and computer assisted intervention-MICCAI 2020: 23rd international conference, Springer, pp 552–561
Xue Y, Tang H, Qiao Z, Gong G, Yin Y, Qian Z, Huang C, Fan W, Huang X (2020) Shape-aware organ segmentation by predicting signed distance maps. In: Proceedings of the AAAI conference on artificial intelligence, pp 12565–12572
Yu L, Wang S, Li X, Fu C, Heng P (2019) Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In: Medical image computing and computer assisted intervention-MICCAI 2019: 22nd international conference, Springer, 2019, pp 605-613
Bernard O, Lalande A et al (2018) Deep learning techniques for automatic MRI cardiac Multi-Structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging 37(11):2514–2525. https://doi.org/10.1109/TMI.2018.2837502
Bai W, Oktay O, Sinclair M, Suzuki H, Rajchl M, Tarroni G, Glocker B, King A, Matthews PM, Rueckert D, (20174) Semi-supervised learning for network-based cardiac MR image segmentation, in. In Medical Image Computing and Computer-Assisted Intervention- MICCAI, (2017) 20th International Conference. Springer, Quebec City, QC, Canada 2017:253–260
Yu L, Wang S, Li X, Fu C, Heng P (2019) Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. Medical image computing and computer assisted intervention-MICCAI 2019: 22nd international conference. Springer, Shenzhen, China, pp 605–613
Verma V, Kawaguchi K, Lamb A, Kannala J, Solin A, Bengio Y, Lopez-Paz D (2022) Interpolation consistency training for semi-supervised learning. Neural Netw 145:90–106. https://doi.org/10.1016/j.neunet.2021.10.008
Acknowledgements
This work was supported by the National Natural Science Foundation of China under grant 12071215 and the Postgraduate Research & Practice Innovation Program of Jiangsu Province [Project number KYCX23 0364].
Author information
Authors and Affiliations
Contributions
Yueyue xiao: Conceptualization, Methodology, Software, Validation, Writing-original draft. Yuan Zou:Data preprocessing, Software, Validation, and analysis. Chunxiao chen: Conceptualization, Writing - review & supervision. Xue Fu Liang Wang, and Jie Yu: Formal analysis and investigation. Huiyu Zhou: review. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Ethical declaration
This article does not contain any studies with human participants and/or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, Y., Chen, C., Fu, X. et al. A novel multi-task semi-supervised medical image segmentation method based on multi-branch cross pseudo supervision. Appl Intell 53, 30343–30358 (2023). https://doi.org/10.1007/s10489-023-05158-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-05158-3