Abstract
Automated image-based plant identification systems are black-boxes, failing to provide an explanation of a classification. Such explanations are seen as being essential by taxonomists and are part of the traditional procedure of plant identification. In this paper, we propose a different method by extracting explicit features from flower images that can be employed to generate explanations. We take the benefit of feature extraction derived from the taxonomic characteristics of plants, with the orchids as an example domain. Feature classifiers were developed using deep neural networks. Two different methods were studied: (1) a separate deep neural network was trained for every individual feature, and (2) a single, multi-label, deep neural network was trained, combining all features. The feature classifiers were tested in predicting 63 orchid species using naive Bayes (NB) and tree-augmented Bayesian networks (TAN). The results show that the accuracy of the feature classifiers is in the range 83-93%. By combining these features using NB and TAN the species can be predicted with an accuracy of 88.9%, which is better than a standard pre-trained deep neural-network architecture, but inferior to a deep learning architecture after fine-tuning of multiple layers. The proposed novel feature extraction method still performs well for identification and is explainable, as opposed to black-box solutions that only aim for the best performance.
Graphical abstract

Similar content being viewed by others
1 Introduction
In traditional plant identification, the determination of a plant’s species follows a fixed procedure, based on the deployment of a combination of taxonomic characteristics, as already initiated by Carl Linnaeus in the eighteenth century Linnaeus [1]. Since that time, after much further improvement, this procedure has evolved into the standard.
In more recent times, a number of fully automated image-based plant identification systems have become available; they can assist both expert and non-expert botanists with the identification process. Example are: LeafSnap Kumar et al. [2] and Plantnet Joly et al. [3]. However, these systems work as a black-box: decisions are provided without explanation. This is not in accordance with standard practice which demands the outcome to be supported and understood in common botanic terms, i.e., being explainable.
To attach a species name to a photographic picture of a plant, a black-box identification system usually employs one of following two approaches. The first approach uses low-level feature extraction such as Moment Invariant Ming-Kuei [4], HSV color, and Histogram of Gradient (HoG), etc. Combining such low-level features may lead to finding the species name as was done by Yang et al. [5], Kho et al. [6], and Lee and Hong [7]. However, features extracted in this way have no botanic meaning, and thus cannot be easily understand by the common user. The second approach uses deep learning such as explored by Liu et al. [8], Ou et al. [9], and Rzanny et al. [10]. Even though deep learning often gives good to superior performance, the features extracted are implicit, hidden, and hard to understand as well.
In this paper, we propose an alternative approach, where the role of explicit, botanic descriptions as features is emphasized. These features are derived from the taxonomic characterization of plants, as used in traditional plant determination. Using these features, the determination of a particular plant as belonging to a particular species can be explained always in terms of the characteristic botanic features used.
The concept of taxonomic feature extraction is quite different from low-level feature extraction. A feature in an image is extracted by a feature classifier, to be combined with other features to determine the species name. Whereas the results of low-level feature extraction are hard to be explained to the user, the results of taxonomic feature extraction are the plant’s characteristics that are already familiar to the user. The difference between feature extraction using – low-level features and taxonomic features – is illustrated in Fig. 1.
In our research we aim to remain close to the traditional approach to plant identification as adopted by botanists, i.e., the explicit, descriptive approach. However, we still wish to profit from the recent advantages in deep learning, and in particular transfer learning, by employing specific successful deep learning neural network architectures and fine-tuning these on new data. Our idea is to employ deep learning for extracting the explicit descriptive features of plants.
Throughout this paper we use the orchids as our example domain of a plant family. There are more than 25,000 orchid species, with some species looking very similar to other orchid species. We only deal with blooming orchids and the explicit features we exploit describe the flower parts of an orchid. The flowers of the orchid are often the most distinguishing part of an orchid. It is often hard to identify a particular orchid species, partly also because there are so many of them, which explains the need for digital support.
The developed method exploits the best of both worlds: the potential of deep neural network architectures in interpreting images, and the easy explainability of plant identification in terms of taxonomic features. However, it is clear that this approach very much relies on how accurate a particular plant feature can be extracted from a digital image.
As far as we are aware of, it is the first time that this combination of methods in computer-based automated plant classification were studied and compared to black-box neural networks. Therefore, the main contributions of this paper are as follows:
-
1.
A number of explainable feature classifiers based on taxonomic features of plant are developed and evaluated.
-
2.
Two different feature classifier methods are compared:
-
(a)
using a separate deep neural network for every individual feature (multi-class classification), and
-
(b)
using a single deep neural network for all features together (multi-label classification).
-
(a)
-
3.
The significance of different feature combinations is determined.
-
4.
The (explicit, taxonomic) feature classifiers are combined in machine learning to predict orchid species and compared to two common deep learning black-box methods.
The remaining part of the paper is organized as follows. Section 2 presents related research in identification of plants using digital images, Section 3 discusses the methods employed in the research. In Section 4, the experiments and their results are presented, which is followed by a discussion in Section 5 and conclusions in Section 6.
2 Related work
In this section, related work in low-level extraction methods, feature extraction and plant identification using deep learning, and other explainable feature extraction methods are discussed.
As already mentioned above, most current automated plant identification approaches are non-descriptive in nature. Some low-level features were used in several studies. Most of the studies make use of color, shape and texture for feature extraction. Sabri et al. [11] extracted HSV color and shape features such as area, perimeter, eccentricity, circularity, etc. on orchid flower image, then applied Support Vector Machine (SVM) for classifying the species. The system achieved the accuracy rate of 82.2%. Shape features employed in this paper are quite understandable. However, those are not invariant to the position of the flower. In this case, the flower image has to be taken from front. In predicting species using low-level features, HSV color and SVM seem to be often used. Another study that applied those features is Andono et al. [12]. They combined HSV color feature and texture feature called GLCM together with SVM, naive Bayes, and k-Nearest Neighbour (k-NN) to classify orchids. Even though applying naive Bayes gives an opportunity to track the decision, however the features extracted in this paper are hard to understand by the common users. Overall, the limitation of the approach mentioned above are: features have neither taxonomic nor biological meaning, systems act as black-boxes, need segmentation to obtain region of interest (ROI) for feature extraction, and the accuracy of the system is not too high.
Nowadays, instead of using low-level features and some machine learning classifiers, deep learning (DL) methods are increasingly used for automated flower recognition. Arwatchananukul et al. [13] built an orchid identification system based on 1500 images from 15 species of orchid flowers. The research used the Inception-v3 as backbone for DL. The performance, up to 98.6% accuracy, was very good, however very uniform images and the same orchids were used multiple times to obtain class balance. Other research concerns work done by Sarachai et al. [14] using homogeneous ensemble of small deep convolutional neural network. Three part of network are used. They claimed that the proposed method can handle the complication of orchids flowers. A study about orchid plant identification using ensemble method also conducted by Ou et al. [9]. An ensemble of three pre-trained model i.e. ResNet50, EfficientNet, and Big Transfer (BiT) were used. The system can achive the accuracy 84.67% which is higher than only using a pre-trained model. In automated flower recognition systems using deep learning, all studies directly process the images into deep learning. In this case, segmentation process did not use. Deep learning approach also often give a high accuracy compared to using low-level features and machine learning classifiers. But, still the systems act as black-boxes.
Research conducted by Farhadi et al. [15] has some similarity to our proposed method. Even though the research was not done in the context of plant or flower identification, this was the first research to explore descriptions of physical objects and subsequently used these in machine learning. Firstly, the low-level features or the base features are extracted from the image. Then, attribute classifiers were trained using the extracted base features. Finally, the features yielded by attribute classifiers were used to learn the object category. According to this research, inferring attributes of objects is key in object recognition. Semantic and discriminative features like parts, shapes and materials can be used for inferring the attributes. This methods does not only allow recognizing objects using an attribute classifier, but also supports describing unfamiliar (new) objects. In other fields, some research for designing explainable feature extraction such as in handwriting recognition Faghihi et al. [16] and medical image analysis Pintelas et al. [17] were found. Faghihi et al. extracts the features by scanning eight defined regions of the image in different direction and counting the number of times they cross a line (from 1 to 4). They claimed that this method can explain how an image was misrecognized. Pintelas et al. propose a new set of explainable features based on mathematical and geometric concepts, such as lines, vertices, contours, and the area size of objects. They carefully selected and created six features category namely “Whole Image”, “Contours numbers”, “Contours Perimeter Size”, “Contours Area Size”, “Contours Vertices Number”, and “Contours Gravity” in order to guarantee explainability.
To the best of our knowledge, we were the first to propose explainable feature extraction using the features commonly employed by taxonomists.
A compact summary of related work is provided in Table 1.
3 Methods
Instead of using deep neural network for directly finding the species name, deep neural network are employed for extracting the taxonomic features of the flower. Figure 2 depicts a diagrammatic summary of our proposed feature extraction method.
A graphical summary of our proposed method. A feature is either extracted by a separate neural network (multi-class classifiers), or all features are extracted together by a single neural network (multi-label classifier); To evaluate whether or not our extracted features can be used to classify an orchid, the extracted features are entered into a classification algorithm that support explainability such as naive Bayes (NB) and Tree-Augmented Bayesian Network (TAN) to determine a species name
3.1 Flower characteristics
Since the work of Linnaeus [1], plants are identified by using a binomial nomenclature, consisting of a genus, followed by the name of the species. For example, Cypripedium is a genus of orchid, the latter being the family of the plant, consisting of 58 species, one of which is Cypripedium montanum (large lady’s slipper). When we speak of ‘species’ in the following, we mean genus followed by species which constitute the ‘name’ of the plant.
In plant determination or identification, the characteristics, taxonomic keys, or features as we call them here, of the plant are always used to determine the species it belongs to. For the purpose of orchid identification from digital photographs, the shape and color of the flower are of primary importance, which explains why in this paper the focus is on flower features. Flowers usually consist of sepals and petals (modified leaves that are the outer sterile whorls of the flower). In orchids, besides sepals and petals, there is also a unique part that can differentiate one orchid flower from others, called labellum or lip. Some of the features of an orchid flower concern shape and structure and are called morphological features. In this research, texture, inflorescence, the number of flowers, labellum characteristics are morphological features. In addition, the color of the flower and the color of labellum are characteristic features. Typical for the features of orchid flowers is that they are almost never unique. Hence, they have to be combined to identify the species of an orchid, and in some cases, even a feature combination does not yield a unique solution.
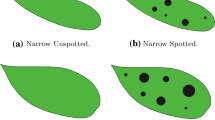
Figure 3 illustrates two morphological features used in this research. All features are discussed one by one in the following:
-
Texture. We only observe texture, abbreviated to T, for the labellum. It is described by the presence of spots or absence of spots (no spots). We call texture a ‘spot’ if there are some regular shapes which have a different color from the base color of the labellum. The spots may be small or large, few or many. We speak of ‘no spots’ in the opposite situation.
-
Inflorescence, or In for short, is the arrangement of flowers on the stem. We group the inflorescence type into 4 groups: panicle, raceme, single or pair, and spike.
-
Number of Flowers, abbreviated to NF, just a count of the number of distinguishable flowers of an orchid, is another relevant characteristic feature.
-
Labellum Characteristic, or LC for short, is the outline of the labellum. We group the labellum characteristics into 4 groups: fringed, lobed, pouched, and simple.
-
Finally, two types of color feature are distinguished. The first one being the Color of Flower, or CF for short, which concerns the color of sepals and petals.
-
The second color used to characterize an orchid is the Color of the Labellum, or CL for brevity.
The complete list of individual features is given in Table 2.
3.2 Multi-class and multi-label classification
To build feature classifiers based on morphological and color features using deep learning, we have two options: (a) using a separate deep neural network for every individual feature (binary and multi-class classification), and (b) using a single deep neural network for all features together (multi-label classification). In this paper, we explore the use of multi-class and multi-label classification to find out which of the two methods is more appropriate to our case. However, first a formal definition of these classification methods is provided.
3.2.1 Multi-class classification
Let database D = {(fi, li)| i = 1, …, N} be a multiset of N tuples or instances (fi, li), with m-tuple \({\textbf{f}}_i=\left({f}_1^i,\dots, {f}_m^i\right)\) composed of values of different features Fj, j = 1, …, m, and \({f}_j^i\in D\left({F}_j\right)\), the (finite or infinite) domain of feature Fj, and \({\textbf{l}}_i=\left({l}_1^i,\dots, {l}_p^i\right)\) being a p-tuple of values of different labels Lk, k = 1, …, p, and \({l}_k^i\in D\left({L}_k\right)\), the finite label domain of label Lk. A classifier C now takes the features of an instance f as input in order to produce label value(s) as output:
A multi-class classifier MC is a function that assigns one label value, usually simply called a class, to a given tuple of feature values, i.e.
with l ∈ D(L1) as p = 1; there is only one set of label values. Multi-class classification concerns classifying an instance into exact one of more than two different label values; if only two label values are distinguished one speaks of binary classification. The complexity of multi-class classification can be similar to single binary classifier Honeine et al. [18].
3.2.2 Multi-label classification
In contrast to multi-class classification, a multi-label classifier ML is a function that assigns one label value to each of the p labels Lj of an instance:
where lj ∈ D(Lj), j = 1, …, p. Thus, in multi-label classification, each instance will be mapped to multiple labels at the same time. For modeling multi-label classification in general, we can use several different methods, which are described below.
a. Problem transformation methods transform the multi-label problem into a set of binary classification problems. There are several methods to transform it into a binary classification problem, for example, binary relevance, classifier chains and label power-set. The simplest method is binary relevance Tsoumakas and Katakis [19]. It works by training independent binary classifiers to predict each label. The independent predictions are then aggregated to form a collection of relevant labels. Classifier chains in Read et al. [20] take a similar approach to binary relevance but explicitly take the associations between labels into account. Label power-set Gupta et al. [21] is well known as the multi-label classification method that has a better performance compared to the other multi-label classification methods. Each unique combination of relevant labels is mapped to a class. This method takes possible correlations between class labels into account. Although this method can perform well, the number of possible unique label combinations grows exponentially, i.e., ∣D(L1) × ⋯ × D(Lp)∣, the cardinality of the Cartesian product of label domains, reducing its usefulness in practical applications.
b. Adapted algorithms try to address the problem in its full form rather than trying to convert the problem to a simpler problem. The algorithms that are most widely used are multi-label k-nearest neighbor, multi-label decision trees, and neural networks Breiman et al. [22]; Bishop [23]. Multi-label k-nearest neighbor uses a binary relevance algorithm, which acts on the labels individually, but instead of applying the standard k-nearest neighbor algorithm directly, it combines with the maximum a posteriori principle. Multi-label decision tree extends the C4.5 decision tree algorithm to allow multiple labels in the leaves, and choose node splits based on a re-defined multi-label entropy function. A type of neural networks that is included in adapted algorithms is back-propagation for multi-label learning (BPMLL).
From the beginning, multi-label classification was inspired by text categorization problems, where each document may belong to several predefined topics simultaneously. Therefore, in this research we investigate whether this method is appropriate for feature classification.
3.3 Deep learning architecture
In the recent past, we already carried out experiments with some well-known pre-trained deep-learning architectures such as VGG-16, ResNet50, InceptionV3, Xception, and NasNet used as color classifiers Apriyanti et al. [24]. The number of parameters used in those deep learning architectures are 138 million, 23.6 million, 54.3 million, 22.8 million, and 22.6 million, respectively Saleem et al. [25]; Radhika et al. [26]. Xception yielded the best performance as color classifier in these experiments. To investigate whether or not Xception would yield the best performance for the other features of the flower, we compared it with the other architectures mentioned above. Compared were the classifier performances for texture and inflorescence of different deep learning architectures with the assumption that sufficient information was obtained to draw conclusions concerning the other four features.
The inputs to the DL networks consisted of images with size 224 × 224 × 3 (where 3 comes from the RGB color coding). For this experiment, we use a pre-trained Xception architecture by freezing the first layer and unfreezing the rest. We added a flatten layer and one dense layer with 256 neurons using the ReLU activation function. We also added a dropout layer with threshold value equal to 0.5.
For multi-class classification the softmax function was chosen as the final layer of the DL architecture, as only a single value for the output was needed, in this case the maximum. The individual class values have to sum up to 1 so that a probabilistic interpretation is allowed. As a consequence, the outputs of a softmax will be all interrelated. If the probability of one class increases, the probabilities of at least one of the other classes will decrease. Each multi-class classifier was trained separately, so there was no dependence between the image features obtained in this way. Figure 4 illustrates the DL architecture used in the multi-class classification scenario.
For multi-label classification, we used deep learning, using the binary relevance method mentioned above (Section 3.2.2(a)), that transforms the values of the features into labels: a positive value is replaced by 1 and a negative value by 0. The number of output neurons was chosen to be equal to the number of labels, where labels were represented using a one-hot encoding so that they could be easier to process in a NN. However, this has no effect on the semantics of the labels, which is kept unchanged. To be able to obtain simultaneous, i.e., non-mutual-exclusive, outputs a sigmoid instead of softmax function was used as the output layer. As the probabilities produced by a sigmoid function are independent there are no mutual constraints amongst the label outputs. The multi-label classification scenario is illustrated in Fig. 5.
Multi-label classification using deep learning resulting in a single neural network with 29 outputs (the complete list of outputs is provided in Table 2)
3.4 Dataset
In our experiments a dataset was used that consisted of orchid flower images with associated ground truth flower characteristics. The images were downloaded from sources such as Flickr, EoL (Encyclopedia of Life), and the Go Botany website, while the morphological characteristics were obtained from descriptions available in the Go Orchid and Go Botany websites.
There are several reasons why this orchid flower dataset is very challenging: flower images have a high variation in size, background, position, and illumination characteristics. The dataset can be freely downloaded from https://doi.org/10.7910/DVN/0HNECY Apriyanti et al. [27].
In the experiments to build feature classifiers, 7156 images were used, split up into 5119 images for training, 1235 image for validations, and 802 images for testing, respectively. The testing set contains new images that were never used for training and validation. The training-data distribution of the features is shown in Fig. 6.
For evaluating the feature classifiers to predict the species using naive Bayes, TAN, InceptionV3 and Xception, we use 6300 images consists of 63 species, and split the images into 5040 images for training, 630 images for validation, and 630 images for testing. The distribution of the orchid species class variable in the training data was balanced in such way that each class consisted of 90 images and associated feature descriptions (where in DL 80 images were used for training, and 10 for validation, and for the Bayesian classifiers all 90 cases per class were used); for the independent test dataset the corresponding number of cases per class was equal to 10.
3.5 Performance evaluation
Common performance evaluation metrics for binary classifiers are true positive rate (TPR) and true negative rate (TNR), in addition to accuracy and the F1 score. TPR represents the number of cases with positive class value that were predicted correctly as being positive, while TNR represents the number of cases with negative class value that are predicted correctly as being negative. For multi-class classification, which we study in this paper, the definitions of these metrics are more elaborate, but still in the same vein; details have been moved to Appendix A.
In addition to an evaluation of the performance of the DL feature classifiers, we also carried out an evaluation of the performance of flower classification methods based on the DL features classifiers and compare the results with the state-of-the-art black-box DL methods which take the image, not the features, as input. Hence, we basically compare DL where vision-based image interpretation is employed, to taxonomic features, still extracted using DL, but interpreted using classifiers. Another way to look as this comparison is as a comparison between whole image, and therefore black-box, interpretation versus image features interpretation. As mentioned in the Introduction, the advantage of using taxonomic features is that classification results can be explained in an understandable manner to the user, as traditionally done by taxonomists.
For the evaluation of the performance using DL methods, we investigated two scenarios, pre-trained without tuning multiple of layers and pre-trained with tuning multiple layers. We chose InceptionV3 and Xception as representative of the blackbox method, therefore we conducted those two scenarios in InceptionV3 and Xception. The first scenario is freezing all layers except the top layer. When we freeze all layers except the top layer (the top layer usually is a classifier), it means that we use the feature extractors and the weight from source domain, to extract the features of images in our target domain. After that, we train the top layer to get the species name. The second scenario is freezing only the first layer. When we freeze only the first layer, it means that we try to tune the network to find better features, because the first layer is expected to provide generic features/low-level features like edges, corners, blobs, etc.
3.6 Software used
Part of the experiments were done using the R language and the R packages bnlearn and gRain Scutari [28]; Højsgaard [29]. In addition, the DL experiments were carried out using the python3 language version 3.7.6 and the tensorflow package Abadi et al. [30]. We also used a high performance computing cluster provided by the University of Twente. For the GPU, we used the NVIDIA QUADRO RTX 6000.
4 Experimental results
4.1 Comparison of different deep learning architectures
Figure 7 shows a comparison between the performance of different deep learning architectures for texture and inflorescence classifiers. It is obvious that the feature classifiers using Xception give the best performance compared to other deep learning architectures, i.e., VGG16, Resnet50, and InceptionV3.
4.2 Multi-class classification results
The results for multi-class classification are summarized in Table 2. In general the feature classifiers yield quite reasonable results. Most of them have a TPR above 0.80. There are only a few features that have a TPR below 0.80 such as: ‘Panicle’ for inflorescence (In), ‘AFew’ for number of flower (NF), ‘Fringed’ for labellum characteristic (LC), ‘GreenYellow’ and ‘PurpleYellow’ for color of flower (CF), ‘GreenRed’, ‘GreenYellow’, and ‘RedYellow’ for color of labellum (CL).
We repeated the experiment three times for each feature classifier using multi-class classification to obtain insight into the variability of their performance. In this case, the variability was produced by different initialisation in deep learning. The results of repeating the experiments are shown in Table 3. From the table it can be concluded that the standard deviation for each classifier is really small – between 0.1% and 1.0% –, meaning that the feature classifiers’ performance appears to be robust.
4.3 Multi-label classification results
For multi-label classification, two methods were explored. First, we used the common multi-label classification method where different levels’ of confidence are used as threshold to classify features. We carried out several experiments using level of confidence, e.g. 0.5, 0.6, and 0.8. However, often this method yielded more than one value for a feature, whereas we aim for a unique classification result. Therefore, another method was used where results, interpreted as a probabilities, were ranked from high to low. The results of this method are shown in Table 2. Overall the TPR is above 80%. The features that have TPR below 80% are exactly the same as in the multi-class classification. Hence, it appears that the results obtained by multi-class and multi-label classification for features are often very similar, but not always, as we will discuss below.
4.4 Comparison of Bayesian feature classifiers and whole image black-box deep learning classifiers
So far we discussed results obtained from DL experiments in extracting features from orchid flower images. However, having obtained these features allows determining the species to which an orchid belongs to, based on those features, in a similar way as a botanist would do, using taxonomic characteristics. For this purpose we trained and validated two different Bayesian network classifiers with the orchid species class variable as a central variable that was linked to the feature variables mentioned above: a naive Bayes classifier Wickramasinghe and Kalutarage [31], and a TAN classifier Zhao et al. [32]. The advantage of a TAN classifier is that dependencies between the feature variables can be captured by the tree structure, which is missing in naive Bayes, where the assumption is made that all features are conditionally independent given the species class variable.
These two Bayesian network classifiers were compared to two DL architectures, InceptionV3 and Xception, where whole images were classified into orchid species by the DL neural networks. The results are shown in Table 4.
5 Discussion
5.1 Flower feature extraction using deep learning
For each of the six orchid flower features we employed in our research it was possible to obtain a reasonably good performance using the Xception architecture, although only after training on the orchid data without which an acceptable result could not be achieved.
We also compared DL for each feature separately (multi-class and binary classification) and multi-label classification, where all features were combined into one neural network. Figure 8 shows a comparison of the performance between multi-class classification and multi-label classification. From the figure, we can conclude that most of all features extracted using multi-class classification outperform multi-label classification, with the exception of ‘AFew’, ‘Many’, and ‘SinglePair’ for number of flowers (NF), ‘Green’ and ‘Red’ for color of labellum (CL), ‘Panicle’ for inflorescence (In), and finally ‘Fringed’ for labellum characteristics (LC), which have a slightly higher performance.
5.2 Feature importance and comparison to black-box deep learning
Using the Bayesian-network classifiers mentioned above, we tried to shed light on which orchid flower feature or combination of features had most impact on the classification performance. The results are summarized in Fig. 9. Concerning the individual feature, the two most important individual features were color of labellum (CL), and color of flower (CF), which also appeared to hold when used as pair, although labellum characteristic (LC) combined with CL achieved similar performance. Adding more features achieves a steady improvement in performance: indeed, using all features gives the best performance. It also is clear that it does not matter much whether naive Bayes or TAN was used as method, i.e. there is no effect of direct interaction between the features.
As Table 4 shows, naive Bayes and TAN yield performance results that are better than the transfer learning without tuning the layers, whereas the best results were obtained by transfer learning with tuning the layers.
5.3 The challenges faced in explainable feature extraction
In this research, data has a main role in obtaining a good performance. We face some challenges during data processing.
5.3.1 Collection of data
We crawled the flower images from multiple websites using API based on the name of the orchid species. We realized that some of the images that we obtained do not contain flower, but only contain bushes, seed, flower painting, etc., so we need to eliminate them from our dataset. This process was time consuming because we had to check it one by one. As we aim to build an explainable classifier, our dataset also includes the flower description of the orchid species. The flower descriptions are based on a taxonomy of plants and we need to integrate flower descriptions from various sources. This was handled by saving the data into a single persistent data storage.
5.3.2 Imbalanced data
This phenomenon also appeared in our dataset. Some species had many images in the dataset, whereas others were represented by only a few images. Imbalance in data is a serious problem in machine learning. We have tried to handle this problem by deep learning using training techniques that take this into account and the orchid class was made balanced (cf. Section 3.4).
5.3.3 Poor description and metadata
Another challenge that we faced in this research is poor flower descriptions and metadata. This problem resulted originally into some wrong annotations of our dataset. The problem was tackled by checking again the description from the literature and visual inspection whether the appearance of the image and the description matched.
5.3.4 Feature selection
Our aim was to extract the features directly from the image. The selected features were derived from the features used by taxonomists in traditional plant identification. The feasibility of extraction of taxonomic features directly from an image varies. Not all taxonomic features can be extracted from a 2D image, and some features play a more important role in plant identification than others. Therefore, feature selection must be done in such a way that the system can distinguish the orchid flowers based on the given set of features.
6 Conclusion
In this paper, a novel flower feature extraction method is proposed based on deep neural networks. We explored the use of a separate deep neural network for every feature (multi-class and binary classification) and a single deep neural network for all features (multi-label classification). Both of the methods can predict the characteristics of the flower very well, with slightly better performance for the separate DL classifiers. The results also show that the proposed explainable feature extraction can be used for orchid identification and yields better results compared to standard pre-trained deep learning. Even though the ultimate performance is inferior to deep learning after fine-tuning of multiple layers, employing explicit taxonomic features offers much better opportunities for explanation, while still providing an acceptable performance.
The features that we selected in this paper do not always suffice to distinguish all types of orchid, therefore inclusion of more features may be needed. Improvement of the performance of automated orchid classification using taxonomic features can also be achieved by improving the performance of feature classifiers, for example by conducting image filtering and image pre-processing.
References
Linnaeus C (1735) Systema naturae, sive regna tria naturae systematice proposita per classes, ordines, genera, & species. Haak, Leiden
Kumar N, Belhumeur PN, Biswas A et al (2012) Leafsnap: A computer vision system for automatic plant species identification. In: Fitzgibbon A, Lazebnik S, Perona P et al (eds) Computer Vision – ECCV 2012. Springer, Berlin Heidelberg, pp 502–516. https://doi.org/10.1007/978-3-642-33709-3_36
Joly A, Bonnet P, Goëau H et al (2016) A look inside the Pl@ntNet experience. Multimed Syst 22(6):751–766. https://doi.org/10.1007/s00530-015-0462-9 URL https://inria.hal.science/hal-01182775
Ming-Kuei H (1962) Visual pattern recognition by moment invariants. IRE Trans Inf Theory 8:179–187
Yang W, Wang S, Zhao X et al (2015) Greenness identification based on hsv decision tree. Inf Process Agric 2(3):149–160. https://doi.org/10.1016/j.inpa.2015.07.003; https://www.sciencedirect.com/science/article/pii/S2214317315000347. Accessed 10 Jan 2023
Kho SJ, Manickam S, Malek S et al (2017) Automated plant identification using artificial neural network and support vector machine. Front Life Sci 10(1):98–107. https://doi.org/10.1080/21553769.2017.1412361
Lee HH, Hong KS (2017) Automatic recognition of flower species in the natural environment. Image Vis Comput 61:98–114. https://doi.org/10.1016/j.imavis.2017.01.013; http://www.sciencedirect.com/science/article/pii/S0262885617300525. Accessed 11 Jan 2023
Liu W, Feng W, Huang M et al (2020) Plant taxonomy in hainan based on deep convolutional neural network and transfer learning. In: 2020 IEEE 19th International Conference on Trust, Security and Privacy in Computing and Communications (TrustCom), pp 1461–1467. https://doi.org/10.1109/TrustCom50675.2020.00197
Ou CH, Hu YN, Jiang DJ et al (2023) An ensemble voting method of pre-trained deep learning models for orchid recognition. In: 2023 IEEE Inter- national Systems Conference (SysCon), pp 1–5. https://doi.org/10.1109/SysCon53073.2023.10131263
Rzanny M, Wittich HC, Mäder P et al (2022) Image-based automated recognition of 31 poaceae species: The most relevant perspectives. Front Plant Sci 12:804140. https://doi.org/10.3389/fpls.2021.804140
Sabri N, Kamarudin MF, Hamzah R et al (2019) Combination of color, shape and texture features for orchid classification. In: 2019 IEEE 9th International Conference on System Engineering and Technology (ICSET), pp 315–319. https://doi.org/10.1109/ICSEngT.2019.8906322
Andono P, Rachmawanto E, Herman N et al (2021) Orchid types classi- fication using supervised learning algorithm based on feature and color extraction. Bull Electr Eng Inform 10(5):2530–2538. https://doi.org/10.11591/eei.v10i5.3118 URL https://beei.org/index.php/EEI/article/view/3118
Arwatchananukul S, Khwunta Kirimasthong K, Aunsri N (2020) A new paphiopedilum orchid database and its recognition using convolutional neural network. Wirel Pers Commun 115:3275–3289
Sarachai W, Bootkrajang J, Chaijaruwanich J et al (2022) Orchid classifica- tion using homogeneous ensemble of small deep convolutional neural net- work. Mach Vis Appl 33(1):17. https://doi.org/10.1007/s00138-021-01267-6
Farhadi A, Endres I, Hoiem D et al. (2009) Describing objects by their attributes. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp 1778–1785. https://doi.org/10.1109/CVPR.2009.5206772
Faghihi F, Cai S, Moustafa A et al. (2022) A nonsynaptic memory based neural network for hand-written digit classification using an explainable feature extraction method. In: Proceedings of the 6th International Conference on Information System and Data Mining. Association for Computing Machinery, New York, NY, USA, ICISDM ‘22, p 69–75. https://doi.org/10.1145/3546157.3546168
Pintelas E, Livieris IE, Pintelas P (2023) Explainable feature extraction and prediction framework for 3d image recognition applied to pneumonia detection. Electronics 12(12):2663. https://doi.org/10.3390/electronics12122663 URL https://www.mdpi.com/2079-9292/12/12/2663
Honeine P, Noumir Z, Richard C (2013) Multiclass classification machines with the complexity of a single binary classifier. Signal Process 93(5):1013–1026. https://doi.org/10.1016/j.sigpro.2012.11.009; https://www.sciencedirect.com/science/article/pii/S0165168412004045. Accessed 9 Jun 2023
Tsoumakas G, Katakis I (2007) Multi-label classification: An overview. Int J Data Warehous Min (IJDWM) 3(3):1–13 URL https://EconPapers.repec.org/RePEc:igg:jdwm00:v:3:y:2007:i:3:p:1-13
Read J, Pfahringer B, Holmes G et al (2011) Classifier chains for multi- label classification. Mach Learn 85(3):333–359. https://doi.org/10.1007/s10994-011-5256-5
Gupta P, Sharma TK, Mehrotra D (2019) Label powerset based multi-label classification for mobile applications. In: Ray K, Sharma TK, Rawat S et al (eds) Soft Computing: Theories and Applications. Springer, Singapore, pp 671–678
Breiman L, Friedman JH, Olshen RA et al (1984) Classification and Regression Trees. Wadsworth and Brooks, Monterey
Bishop C (2005) Pattern Recognition and Machine Learning. Springer
Apriyanti DH, Spreeuwers LJ, Lucas PJF et al (2021) Automated color detection in orchids using color labels and deep learning. PLoS One 16:1–27. https://doi.org/10.1371/JOURNAL.PONE.0259036
Saleem MH, Potgieter J, Arif KM (2020) Plant disease classification: A com- parative evaluation of convolutional neural networks and deep learning optimizers. Plants (Basel) 9(10):1319
Radhika K, Devika K, Aswathi T et al (2020) Performance Analysis of NASNet on Unconstrained Ear Recognition. Springer International Publishing, Cham, pp 57–82. https://doi.org/10.1007/978-3-030-33820-6_3
Apriyanti D, Spreeuwers L, Lucas P, et al. (2020) Orchid Flowers Dataset. https://doi.org/10.7910/DVN/0HNECY
Scutari M (2010) Learning Bayesian networks with the bnlearn R package. J Stat Softw 35(3):1–22. http://cran.r-project.org/web/packages/bnlearn/bnlearn.pdf. Accessed 5 Feb 2023
Højsgaard S (2012) Graphical independence networks with the gRain package for R. J Stat Softw 46(10):1–26. https://doi.org/10.18637/jss.v046.i10; https://www.jstatsoft.org/v46/i10/. Accessed 13 Feb 2023
Abadi M, Agarwal A, Barham P et al (2015) TensorFlow: Large-scale machine learning on heterogeneous systems. https://www.tensorflow.org/, soft-ware available from tensorflow.org. Accessed 4 Jan 2023
Wickramasinghe I, Kalutarage H (2021) Naive bayes: applications, varia- tions and vulnerabilities: a review of literature with code snippets for implementation. Soft Comput 25(3):2277–2293. https://doi.org/10.1007/s00500-020-05297-6
Zhao J, Liu J, Sun Y et al (2011) Tree augmented näıve possibilistic network classifier. In: 2011 Eighth International Conference on Fuzzy Systems and Knowledge Discovery (FSKD), pp 1065–1069. https://doi.org/10.1109/FSKD.2011.6019738
Acknowledgments
The research described in this paper was funded by Research and Innovation in Science and Technology Project (RISET-Pro) of the Ministry of Research, Technology, and Higher Education of Republic Indonesia (World Bank Loan No.8245-ID) and supported by Research and Innovation Agency (BRIN), Republic of Indonesia. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A Classifier evaluation metrics
Appendix A Classifier evaluation metrics
A common method used to evaluate the performance of a classifier (binary or multi-class) is to use summary statistics based on the n × n confusion matrix C (by definition a square matrix), as shown in Table 5, where xij, 1 ≤ i, j ≤ n, stands for the number of cases where the actual value vi is predicted as vj by the classifier. For the binary case, i.e., n = 2, it is common to define the true positive rate (TPR) as
where x11 are known as the true positives (TPs), and x12 as the false negatives (FNs). Similarly, a true negative rate (TNR) is defined as:
where x22 are the true negatives (TNs) and x21 the false positives (FPs). Together these measures offer a good summary of the performance of a classifier.
However, in the case of a multi-class classifier (i.e., n > 2) it is no longer possible to make a distinction between positive and negative and we have instead to redefine the measures TPR and TNR for the individual class values. Let \({n}_i=\sum\nolimits_{j=1}^n{x}_{i,j}\) be the total number of cases with actual value vi and predicted value vj, 1 ≤ j ≤ n, i.e., the sum of row i of the confusion matrix C. The TPR for class value vi, denoted TPRi is defined as follows:
The TPR is also known as the recall, in particular in information retrieval literature, but also sometimes in machine learning.
The definition of the true negative rate for class value vi is more complicated. First we have to exclude those elements from the confusion matrix regarding the predicted and actual value vi. All the other elements are part of the true negatives. Second, we have to compute the false positives, which is done by summing together the values of column i with the exception of xii (the TPs). This results in the following formula:
The denominator of eq. (A4) can be simplified by realizing that it represents the sum of elements in the part of the confusion matrix after removing row i:
resulting in the following definition of the TNR:
A common performance metric to describe the global performance of multi-class classifier across different class values is accuracy, normally only used when the data has class balance (possibly after resolving class imbalance), which is defined as the sum of the elements along the diagonal of the confusion matrix C, divided by the sum of all elements of C:
The last performance measure we will use is called the \({\textbf{F}}_{\textbf{1}}^{\boldsymbol{i}}\) score for class value i, that combines more of the information from the confusion matrix than accuracy and thus is normally seen as a better measure than accuracy for imbalanced data. It is defined as follows:
where recalli, which is identical to the TPRi, is defined above; precisioni is defined as follows:
where TPi = xii and \({\mathrm{FP}}_i\sum\nolimits_{\underset{j\ne i}{1\le j\le n,}}{x}_{ji}\).
As an example, consider the confusion matrix shown in Table 6.
We first compute the TPR (or recall) for a, b, and c:
Subsequently, the TNRs are computed:
The (overall) accuracy of the classifier is equal to:
The \({\mathrm{F}}_1^i\) scores are being computed next:
where precisiona = 20/(20 + 1 + 4) = 0.8,
where precisionb = 25/(25 + 3 + 3) = 0.8,
where precisionc = 11/(11 + 1 + 2) ≈ 0.79.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Apriyanti, D.H., Spreeuwers, L.J. & Lucas, P.J. Deep neural networks for explainable feature extraction in orchid identification. Appl Intell 53, 26270–26285 (2023). https://doi.org/10.1007/s10489-023-04880-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-04880-2