Abstract
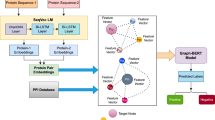
Knowledge of protein-protein interactions (PPI) is essential for studying protein functions and understanding the biological processes. Previously, most of the works on PPI in the BioNLP domain rely solely on textual data. With the availability of different information (structure, sequence, gene ontology) about proteins, researchers have started to use other details with textual data to predict PPI more accurately. This paper reports the first attempt in integrating gene ontology(GO)-based information with the features extracted from other two modalities of proteins namely 3D structure and existing textual information. Existing two popular text-based benchmark PPI corpora, i.e., BioInfer and HRPD50 are first extended to integrate with the structure and GO-based information. Finally, some deep learning-based techniques are employed to extract features from three modalities and those are concatenated for final prediction of protein interaction. The experimentation on generated multi-modal datasets illustrates that the proposed deep multi-modal framework outperforms the baselines (uni-modal, bi-modal and multi-modal) and state-of-the-art methods.





Similar content being viewed by others
Data availibility
The data that support the findings of this study are openly available at https://github.com/sduttap16/MM_PPI_NLP.
References
Airola, A., Pyysalo, S., Björne, J., Pahikkala, T., Ginter, F., & Salakoski, T. (2008). All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinformatics, 9(11), S2.
Airola, A., Pyysalo, S., Björne, J., Pahikkala, T., Ginter, F., Salakoski, T (2008). A graph kernel for protein-protein interaction extraction. In: Proceedings of the workshop on current trends in biomedical natural language processing, pp. 1–9. Association for Computational Linguistics.
Alizadehsani, R., Roshanzamir, M., Hussain, S., Khosravi, A., Koohestani, A., Zangooei, M.H., Abdar, M., Beykikhoshk, A., Shoeibi, A., Zare, A., et al. (2021). Handling of uncertainty in medical data using machine learning and probability theory techniques: A review of 30 years (1991–2020). Annals of Operations Research pp. 1–42.
Asada, M., Miwa, M., Sasaki, Y. (2018). Enhancing drug-drug interaction extraction from texts by molecular structure information. arXiv preprint arXiv:1805.05593.
Bahdanau, D., Cho, K., Bengio, Y. (2014). Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473.
Biro, J. (2006). Amino acid size, charge, hydropathy indices and matrices for protein structure analysis. Theoretical Biology and Medical Modelling, 3(1), 1–12.
Bunescu, R., Ge, R., Kate, R. J., Marcotte, E. M., Mooney, R. J., Ramani, A. K., & Wong, Y. W. (2005). Comparative experiments on learning information extractors for proteins and their interactions. Artificial Intelligence in Medicine, 33(2), 139–155.
Chen, J., Lim, C.P., Tan, K.H., Kumar, A. (2021). Artificial intelligence-based human-centric decision support framework: an application to predictive maintenance in asset management under pandemic environments. Annals of Operation Research.
Choi, S. P. (2018). Extraction of protein-protein interactions (ppis) from the literature by deep convolutional neural networks with various feature embeddings. Journal of Information Science, 44(1), 60–73.
Choi, S.P., Myaeng, S.H. (2010). Simplicity is better: revisiting single kernel ppi extraction. In: Proceedings of the 23rd international conference on computational linguistics, pp. 206–214. Association for Computational Linguistics.
Consortium, G. O., Consortium. (2006). The gene ontology (go) project in 2006. Nucleic Acids Research, 34(suppl–1), D322–D326.
Devlin, J., Chang, M.W., Lee, K., Toutanova, K. (2018). Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
Dutta, P., & Saha, S. (2017). Fusion of expression values and protein interaction information using multi-objective optimization for improving gene clustering. Computers in Biology and Medicine, 89, 31–43.
Dutta, P., Saha, S., & Gulati, S. (2019). Graph-based hub gene selection technique using protein interaction information: Application to sample classification. IEEE Journal of Biomedical and Health Informatics, 23(6), 2670–2676.
Ekbal, A., Saha, S., Bhattacharyya, P., et al. (2016). A deep learning architecture for protein-protein interaction article identification. In: 2016 23rd international conference on pattern recognition (ICPR), pp. 3128–3133. IEEE.
Erkan, G., Ozgur, A., Radev, D.R. (2007). Semi-supervised classification for extracting protein interaction sentences using dependency parsing. In: Proceedings of the 2007 joint conference on empirical methods in natural language processing and computational natural language learning (EMNLP-CoNLL).
He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778.
Hegde, V., Zadeh, R. (2016). Fusionnet: 3d object classification using multiple data representations. arXiv preprint arXiv:1607.05695.
Hsieh, Y.L., Chang, Y.C., Chang, N.W., Hsu, W.L. (2017). Identifying protein-protein interactions in biomedical literature using recurrent neural networks with long short-term memory. In: Proceedings of the eighth international joint conference on natural language processing (volume 2: short papers), pp. 240–245.
Hua, L., Quan, C. (2016). A shortest dependency path based convolutional neural network for protein-protein relation extraction. BioMed Research International 2016.
Huang, M., Zhu, X., Hao, Y., Payan, D. G., Qu, K., & Li, M. (2004). Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics, 20(18), 3604–3612.
Jin, M., Bahadori, M.T., Colak, A., Bhatia, P., Celikkaya, B., Bhakta, R., Senthivel, S., Khalilia, M., Navarro, D., Zhang, B., et al. (2018). Improving hospital mortality prediction with medical named entities and multimodal learning. arXiv preprint arXiv:1811.12276
Khare, R., Leaman, R., Lu, Z. (2014). Accessing biomedical literature in the current information landscape. In: Biomedical Literature Mining, pp. 11–31. Springer.
Kocheturov, A., Pardalos, P. M., & Karakitsiou, A. (2019). Massive datasets and machine learning for computational biomedicine: trends and challenges. Annals of Operations Research, 276(1), 5–34.
Kulmanov, M., Khan, M. A., & Hoehndorf, R. (2017). Deepgo: Predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics, 34(4), 660–668.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2019). BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz682
Li, L., Guo, R., Jiang, Z., & Huang, D. (2015). An approach to improve kernel-based protein-protein interaction extraction by learning from large-scale network data. Methods, 83, 44–50.
Liu, S., Vicente, L.N. (2021). The stochastic multi-gradient algorithm for multi-objective optimization and its application to supervised machine learning. Annals of Operations Research pp. 1–30.
Miwa, M., Sætre, R., Miyao, Y., & Tsujii, J. (2009). Protein-protein interaction extraction by leveraging multiple kernels and parsers. International Journal of Medical Informatics, 78(12), e39–e46.
Moschitti, A. (2006). Making tree kernels practical for natural language learning. In: 11th conference of the European Chapter of the Association for Computational Linguistics.
Ono, T., Hishigaki, H., Tanigami, A., & Takagi, T. (2021). Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics, 17(2), 155–161.
Palaga, P. (2009). Extracting relations from biomedical texts using syntactic information (p. 138). Mémoire de DEA: Technische Universität Berlin.
Peissig, P. L., Rasmussen, L. V., Berg, R. L., Linneman, J. G., McCarty, C. A., Waudby, C., Chen, L., Denny, J. C., Wilke, R. A., Pathak, J., et al. (2012). Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. Journal of the American Medical Informatics Association, 19(2), 225–234.
Peng, Y., Lu, Z. (2017). Deep learning for extracting protein-protein interactions from biomedical literature. arXiv preprint arXiv:1706.01556 .
Pesquita, C., Faria, D., Falcao, A. O., Lord, P., & Couto, F. M. (2009). Semantic similarity in biomedical ontologies. PLoS Computational Biology, 5(7), e1000443.
Poria, S., Hazarika, D., Majumder, N., Naik, G., Cambria, E., Mihalcea, R. (2018). Meld: A multimodal multi-party dataset for emotion recognition in conversations. arXiv preprint arXiv:1810.02508.
Pyysalo, S., Airola, A., Heimonen, J., Björne, J., Ginter, F., Salakoski, T. (2008). Comparative analysis of five protein-protein interaction corpora. In: BMC bioinformatics, vol. 9, p. S6. BioMed Central.
Qian, L., & Zhou, G. (2012). Tree kernel-based protein-protein interaction extraction from biomedical literature. Journal of Biomedical Informatics, 45(3), 535–543.
Qiao, Z., Wu, X., Ge, S., & Fan, W. (2019). Mnn: Multimodal attentional neural networks for diagnosis prediction. Extraction, 1, A1.
Qureshi, S.A., Dias, G., Hasanuzzaman, M., Saha, S. (2020). Improving depression level estimation by concurrently learning emotion intensity. IEEE Computational Intelligence Magazine.
Qureshi, S. A., Saha, S., Hasanuzzaman, M., & Dias, G. (2019). Multitask representation learning for multimodal estimation of depression level. IEEE Intelligent Systems, 34(5), 45–52.
Sabour, S., Frosst, N., Hinton, G.E. (2017). Dynamic routing between capsules. In: Advances in neural information processing systems, pp. 3856–3866.
Sætre, R., Sagae, K., Tsujii, J. (2007). Syntactic features for protein-protein interaction extraction. LBM (Short Papers) 319.
Saha, S., et al. (2020). Amalgamation of protein sequence, structure and textual information for improving protein-protein interaction identification. In: Proceedings of the 58th annual meeting of the association for computational linguistics, pp. 6396–6407.
Smaili, F. Z., Gao, X., & Hoehndorf, R. (2019). Opa2vec: Combining formal and informal content of biomedical ontologies to improve similarity-based prediction. Bioinformatics, 35(12), 2133–2140.
Tikk, D., Thomas, P., Palaga, P., Hakenberg, J., & Leser, U. (2010). A comprehensive benchmark of kernel methods to extract protein-protein interactions from literature. PLoS Computational Biology, 6(7), e1000837.
Ünlü, R., & Xanthopoulos, P. (2019). A weighted framework for unsupervised ensemble learning based on internal quality measures. Annals of Operations Research, 276(1), 229–247.
Van Landeghem, S., Saeys, Y., De Baets, B., Van de Peer, Y (2008). Extracting protein-protein interactions from text using rich feature vectors and feature selection. In: 3rd International symposium on Semantic Mining in Biomedicine (SMBM 2008), pp. 77–84. Turku Centre for Computer Sciences (TUCS).
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, L., Polosukhin, I. (2017). Attention is all you need. arXiv preprint arXiv:1706.03762.
Yadav, S., Ekbal, A., Saha, S., Kumar, A., & Bhattacharyya, P. (2019). Feature assisted stacked attentive shortest dependency path based bi-lstm model for protein-protein interaction. Knowledge-Based Systems, 166, 18–29.
Zhang, S., Wang, X., Liu, A., Zhao, C., Wan, J., Escalera, S., Shi, H., Wang, Z., Li, S.Z (2019). A dataset and benchmark for large-scale multi-modal face anti-spoofing. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 919–928.
Zhao, Z., Yang, Z., Lin, H., Wang, J., & Gao, S. (2016). A protein-protein interaction extraction approach based on deep neural network. International Journal of Data Mining and Bioinformatics, 15(2), 145–164.
Acknowledgements
Kanchan Jha and Dr. Sriparna Saha would like to acknowledge the support of Science and Engineering Research Board (SERB) of Department of Science and Technology India to carry out this research.
Author information
Authors and Affiliations
Ethics declarations
Conflict of interest
All the authors declare that they do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jha, K., Saha, S. & Dutta, P. Incorporation of gene ontology in identification of protein interactions from biomedical corpus: a multi-modal approach. Ann Oper Res (2022). https://doi.org/10.1007/s10479-022-04527-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s10479-022-04527-4