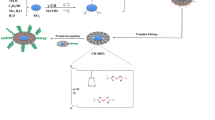
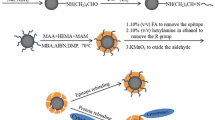
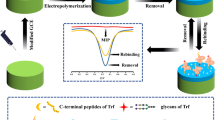
Abstract
The C-terminal epitope imprinted polymers on the silica were prepared by reversible addition-fragmentation chain transfer (RAFT) strategy with C-terminal nonapeptide of cytochrome c (Cyt c) as the template. 4-cyano-4-(phenylcarbonothioylthio) phetanoic acid was immobilized on the silica as a chain transfer agent to regulate the polymerization of the imprinted layer to enhance the recognition performance of the imprinted materials for the peptide and target protein. Additionally, zinc methacrylate was used as a functional monomer to form the imprinted sites through the synergistic effect of Zn2+ chelating, hydrogen bonding and electrostatic attraction, which further improved the recognition between polymers and protein. The molecularly imprinted polymers prepared with RAFT strategy show good recognition for the epitope peptide and Cyt c with imprinting factors of 7.96 and 6.17, respectively, which is higher than those prepared without RAFT strategy. Furthermore, the imprinted polymers have a good binding capacity and imprinting factor for Cyt c in multi-proteins selective recognition and have good reusability. The performance of Cyt c recognition in bovine serum by the epitope imprinted polymers was calculated by HPLC and the results demonstrated the well selectivity and potential application for Cyt c recognition in biological sample.
Graphical abstract










Similar content being viewed by others
References
Cheong WJ, Yang SH, Ali F (2013) Molecular imprinted polymers for separation science: a review of reviews. J Sep Sci 36(3):609–628. https://doi.org/10.1002/jssc.201200784
Fu GQ, He HY, Chai ZH, Chen HC, Kong JA, Wang Y, Jiang YZ (2011) Enhanced lysozyme imprinting over nanoparticles functionalized with carboxyl groups for noncovalent template sorption. Anal Chem 83(4):1431–1436. https://doi.org/10.1021/ac1029924
Guo T, Deng QL, Fang GZ, Liu CC, Huang X, Wang S (2015) Molecularly imprinted upconversion nanoparticles for highly selective and sensitive sensing of Cytochrome c. Biosensors Bioelectron 74:498–503. https://doi.org/10.1016/j.bios.2015.06.058
Niu H, Yang YQ, Zhang HQ (2015) Efficient one-pot synthesis of hydrophilic and fluorescent molecularly imprinted polymer nanoparticles for direct drug quantification in real biological samples. Biosensors Bioelectron 74:440–446. https://doi.org/10.1016/j.bios.2015.06.071
Zhang ZL, Zhang XD, Niu DC, Li YS, Shi JL (2017) Large-pore, silica particles with antibody-like, biorecognition sites for efficient protein separation. J Mater Chem B 5(22):4214–4220. https://doi.org/10.1039/c7tb00886d
Wulff G, Gross T, Schonfeld R (1997) Enzyme models based on molecularly imprinted polymers with strong esterase activity. Angew Chem Int Ed Engl 36(18):1962–1964. https://doi.org/10.1002/anie.199719621
Xu SX, Wang LS, Liu Z (2021) Molecularly imprinted polymer nanoparticles: an emerging versatile platform for cancer therapy. Angew Chem Int Ed 60(8):3858–3869. https://doi.org/10.1002/anie.202005309
Yang Q, Li JH, Wang XY, Peng HL, Xiong H, Chen LX (2018) Strategies of molecular imprinting-based fluorescence sensors for chemical and biological analysis. Biosens Bioelectron 112:54–71. https://doi.org/10.1016/j.bios.2018.04.028
Ye L, Haupt K (2004) Molecularly imprinted polymers as antibody and receptor mimics for assays, sensors and drug discovery. Anal Bioanal Chem 378(8):1887–1897. https://doi.org/10.1007/s00216-003-2450-8
Zhang N, Xu YR, Li ZL, Yan CR, Mei K, Ding ML, Ding SC, Guan P, Qian LW, Du CB, Hu XL (2019) Molecularly imprinted materials for selective biological recognition. Macromol Rapid Commun 40(17):21. https://doi.org/10.1002/marc.201900096
Gu ZK, Dong YR, Xu SX, Wang LS, Liu Z (2021) Molecularly imprinted polymer-based smart prodrug delivery system for specific targeting, prolonged retention, and tumor microenvironment-triggered release. Angew Chem Int Ed 60(5):2663–2667. https://doi.org/10.1002/anie.202012956
Zahedi P, Ziaee M, Abdouss M, Farazin A, Mizaikoff B (2016) Biomacromolecule template-based molecularly imprinted polymers with an emphasis on their synthesis strategies: a review. Polym Adv Technol 27(9):1124–1142. https://doi.org/10.1002/pat.3754
Hua ZD, Zhou S, Zhao MP (2009) Fabrication of a surface imprinted hydrogel shell over silica microspheres using bovine serum albumin as a model protein template. Biosens Bioelectron 25(3):615–622. https://doi.org/10.1016/j.bios.2009.01.027
Lu Y, Li CX, Liu XH, Huang WQ (2002) Molecular recognition through the exact placement of functional groups on non-covalent molecularly imprinted polymers. J Chromatogr A 950(1–2):89–97. https://doi.org/10.1016/s0021-9673(02)00058-4
Mosbach K, Haupt K (1998) Some new developments and challenges in non-covalent molecular imprinting technology. J Mol Recognit 11(1–6):62–68. https://doi.org/10.1002/(sici)1099-1352(199812)11:1/6%3c62::Aid-jmr391%3e3.0.Co;2-5
Hsieh RY, Tsai HA, Syu MJ (2006) Designing a molecularly imprinted polymer as an artificial receptor for the specific recognition of creatinine in serums. Biomaterials 27(9):2083–2089. https://doi.org/10.1016/j.biomaterials.2005.09.024
Turner NW, Jeans CW, Brain KR, Allender CJ, Hlady V, Britt DW (2006) From 3D to 2D: a review of the molecular imprinting of proteins. Biotechnol Progr 22(6):1474–1489. https://doi.org/10.1021/bp060122g
Ge Y, Turner APF (2008) Too large to fit? Recent developments in macromolecular imprinting. Trends Biotechnol 26(4):218–224. https://doi.org/10.1016/j.tibtech.2008.01.001
Li SW, Yang KG, Liu JX, Jiang B, Zhang LH, Zhang YK (2015) Surface-imprinted nanoparticles prepared with a his-tag-anchored epitope as the template. Anal Chem 87(9):4617–4620. https://doi.org/10.1021/ac5047246
Wu G, Li JY, Qu X, Zhang YX, Hong H, Liu CS (2015) Template size matched film thickness for effectively in situ surface imprinting: a model study of glycoprotein imprints. Rsc Adv 5(58):47010–47021. https://doi.org/10.1039/c5ra06454f
Yang KG, Zhang LH, Liang Z, Zhang YK (2012) Protein-imprinted materials: rational design, application and challenges. Anal Bioanal Chem 403(8):2173–2183. https://doi.org/10.1007/s00216-012-5840-y
Liu JX, Deng QL, Tao DY, Yang KG, Zhang LH, Liang Z, Zhang YK (2014) Preparation of protein imprinted materials by hierarchical imprinting techniques and application in selective depletion of albumin from human serum. Sci Rep 4:6. https://doi.org/10.1038/srep05487
Moczko E, Guerreiro A, Caceres C, Piletska E, Sellergren B, Piletsky SA (2019) Epitope approach in molecular imprinting of antibodies. J Chromatogr B 1124:1–6. https://doi.org/10.1016/j.jchromb.2019.05.024
Nothling MD, Fu Q, Reyhani A, Allison-Logan S, Jung K, Zhu J, Kamigaito M, Boyer C, Qiao GG (2020) Progress and perspectives beyond traditional RAFT polymerization. Adv Sci 7(20):12. https://doi.org/10.1002/advs.202001656
Yang KG, Li SW, Liu LK, Chen YW, Zhou W, Pei JQ, Liang Z, Zhang LH, Zhang YK (2019) Epitope imprinting technology: progress, applications, and perspectives toward artificial antibodies. Adv Mater 31(50):17. https://doi.org/10.1002/adma.201902048
Yang KG, Liu JX, Li SW, Li QR, Wu Q, Zhou Y, Zhao Q, Deng N, Liang Z, Zhang LH, Zhang YK (2014) Epitope imprinted polyethersulfone beads by self-assembly for target protein capture from the plasma proteome. Chem Commun 50(67):9521–9524. https://doi.org/10.1039/c4cc03428g
Yang YQ, He XW, Wang YZ, Li WY, Zhang YK (2014) Epitope imprinted polymer coating CdTe quantum dots for specific recognition and direct fluorescent quantification of the target protein bovine serum albumin. Biosens Bioelectron 54:266–272. https://doi.org/10.1016/j.bios.2013.11.004
Xing R, Ma Y, Wang Y, Wen Y, Liu Z (2019) Specific recognition of proteins and peptides via controllable oriented surface imprinting of boronate affinity-anchored epitopes. Chem Sci 10(6):1831–1835. https://doi.org/10.1039/c8sc04169e
Moad G, Rizzardo E, Thang SH (2006) Living radical polymerization by the RAFT process—a first update. Aust J Chem 59(10):669–692. https://doi.org/10.1071/ch06250
Xiao YH, Xiao R, Tang J, Zhu QK, Li XM, Xiong Y, Wu XW (2017) Preparation and adsorption properties of molecularly imprinted polymer via RAFT precipitation polymerization for selective removal of aristolochic acid I. Talanta 162:415–422. https://doi.org/10.1016/j.talanta.2016.10.014
Montagna V, Haupt K, Gonzato C (2020) RAFT coupling chemistry: a general approach for post-functionalizing molecularly imprinted polymers synthesized by radical polymerization. Polyme Chem 11(5):1055–1061. https://doi.org/10.1039/c9py01629e
Azizi A, Shahhoseini F, Bottaro CS (2020) Magnetic molecularly imprinted polymers prepared by reversible addition fragmentation chain transfer polymerization for dispersive solid phase extraction of polycyclic aromatic hydrocarbons in water. J Chromatogr A. https://doi.org/10.1016/j.chroma.2019.460534
Kuscuoglu CK, Guner H, Soylemez MA, Guven O, Barsbay M (2019) A smartphone-based colorimetric PET sensor platform with molecular recognition via thermally initiated RAFT-mediated graft copolymerization. Sens Actuators B 296:12. https://doi.org/10.1016/j.snb.2019.126653
Zhou L, Wang Y, Xing R, Chen J, Liu J, Li W, Liu Z (2019) Orthogonal dual molecularly imprinted polymer-based plasmonic immunosandwich assay: A double characteristic recognition strategy for specific detection of glycoproteins. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2019.111729
Min Y, Jiang B, Wu C, Xia SM, Zhang XD, Liang Z, Zhang LH, Zhang YK (2014) 1.9 mu m superficially porous packing material with radially oriented pores and tailored pore size for ultra-fast separation of small molecules and biomolecules. J Chromatogr A 1356:148–156. https://doi.org/10.1016/j.chroma.2014.06.049
Li QR, Yang KG, Liang Y, Jiang B, Liu JX, Zhang LH, Liang Z, Zhang YK (2014) Surface protein imprinted core-shell particles for high selective lysozyme recognition prepared by reversible addition-fragmentation chain transfer strategy. Acs Appl Mater Interfaces 6(24):21954–21960. https://doi.org/10.1021/am5072783
Okan M, Sari E, Duman M (2017) Molecularly imprinted polymer based micromechanical cantilever sensor system for the selective determination of ciprofloxacin. Biosens Bioelectron 88:258–264. https://doi.org/10.1016/j.bios.2016.08.047
Okan M, Duman M (2018) Functional polymeric nanoparticle decorated microcantilever sensor for specific detection of erythromycin. Sens Actuators B 256:325–333. https://doi.org/10.1016/j.snb.2017.10.098
Qin YP, Wang HY, He XW, Li WY, Zhang YK (2018) Metal chelation dual-template epitope imprinting polymer via distillation-precipitation polymerization for recognition of porcine serum albumin. Talanta 185:620–627. https://doi.org/10.1016/j.talanta.2018.03.082
Moad G, Rizzardo E, Thang SH (2011) End-functional polymers, thiocarbonylthio group removal/transformation and reversible addition-fragmentation-chain transfer (RAFT) polymerization. Polym Int 60(1):9–25. https://doi.org/10.1002/pi.2988
Gemici H, Legge TM, Whittaker M, Monteiro MJ, Perrier S (2007) Original approach to multiblock copolymers via reversible addition-fragmentation chain transfer polymerization. J Polym Sci Part A-1 Polym Chem 45(11):2334–2340. https://doi.org/10.1002/pola.21985
Meng Y, Wei Z, Lu YL, Zhang LQ (2012) Structure, morphology, and mechanical properties of polysiloxane elastomer composites prepared by in situ polymerization of zinc dimethacrylate. Express Polym Lett 6(11):882–894. https://doi.org/10.3144/expresspolymlett.2012.94
Zhao Q-Y, Zhao H-T, Yang X, Zhang H, Dong A-J, Wang J, Li B (2018) Selective recognition and fast enrichment of anthocyanins by dummy molecularly imprinted magnetic nanoparticles. J Chromatogr A 1572:9–19. https://doi.org/10.1016/j.chroma.2018.08.029
Amornchaiyapitak C, Taweepreda W, Tangboriboonrat P (2008) Modification of epoxidised natural rubber film surface by polymerisation of methyl methacrylate. Eur Polym J 44(6):1782–1788. https://doi.org/10.1016/j.eurpolymj.2008.03.002
Du CB, Hu XL, Guan P, Gao XM, Song RY, Li J, Qian LW, Zhang N, Guo LX (2016) Preparation of surface-imprinted microspheres effectively controlled by orientated template immobilization using highly cross-linked raspberry-like microspheres for the selective recognition of an immunostimulating peptide. J Mater Chem B 4(8):1510–1519. https://doi.org/10.1039/c5tb02633d
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21804099).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, M., Li, X., Zhang, H. et al. Preparation of C-Terminal Epitope Imprinted Particles Via Reversible Addition-Fragmentation Chain Transfer Polymerization and Zn2+ Chelating Strategy: Selective Recognition of Cytochrome c. Chromatographia 85, 743–754 (2022). https://doi.org/10.1007/s10337-022-04180-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-022-04180-w