Abstract
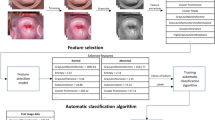
Cervical cancer is the most common cancer among women worldwide. The diagnosis and classification of cancer are extremely important, as it influences the optimal treatment and length of survival. The objective was to develop and validate a diagnosis system based on convolutional neural networks (CNN) that identifies cervical malignancies and provides diagnostic interpretability. A total of 8496 labeled histology images were extracted from 229 cervical specimens (cervical squamous cell carcinoma, SCC, n = 37; cervical adenocarcinoma, AC, n = 8; nonmalignant cervical tissues, n = 184). AlexNet, VGG-19, Xception, and ResNet-50 with five-fold cross-validation were constructed to distinguish cervical cancer images from nonmalignant images. The performance of CNNs was quantified in terms of accuracy, precision, recall, and the area under the receiver operating curve (AUC). Six pathologists were recruited to make a comparison with the performance of CNNs. Guided Backpropagation and Gradient-weighted Class Activation Mapping (Grad-CAM) were deployed to highlight the area of high malignant probability. The Xception model had excellent performance in identifying cervical SCC and AC in test sets. For cervical SCC, AUC was 0.98 (internal validation) and 0.974 (external validation). For cervical AC, AUC was 0.966 (internal validation) and 0.958 (external validation). The performance of CNNs falls between experienced and inexperienced pathologists. Grad-CAM and Guided Gard-CAM ensured diagnoses interpretability by highlighting morphological features of malignant changes. CNN is efficient for histological image classification tasks of distinguishing cervical malignancies from benign tissues and could highlight the specific areas of concern. All these findings suggest that CNNs could serve as a diagnostic tool to aid pathologic diagnosis.





Similar content being viewed by others
References
Arbyn, M., et al., Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health, 2020. 8(2): p. e191-e203.
Gien, L.T., M.C. Beauchemin, and G. Thomas, Adenocarcinoma: a unique cervical cancer. Gynecol Oncol, 2010. 116(1): p. 140-6.
Siegel, R.L., et al., Cancer Statistics, 2021. CA Cancer J Clin, 2021. 71(1): p. 7-33.
Wu, S.Y., E.Y. Huang, and H. Lin, Optimal treatments for cervical adenocarcinoma. Am J Cancer Res, 2019. 9(6): p. 1224-1234.
Sun, H., et al., Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms. IEEE J Biomed Health Inform, 2020. 24(6): p. 1664-1676.
Albayrak, A., et al., A whole-slide image grading benchmark and tissue classification for cervical cancer precursor lesions with inter-observer variability. Med Biol Eng Comput, 2021. 59(7-8): p. 1545-1561.
Pouliakis, A., et al., Using classification and regression trees, liquid-based cytology and nuclear morphometry for the discrimination of endometrial lesions. Diagn Cytopathol, 2014. 42(7): p. 582-91.
LeCun, Y., Y. Bengio, and G. Hinton, Deep learning. Nature, 2015. 521(7553): p. 436-44.
Ehteshami Bejnordi, B., et al., Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama, 2017. 318(22): p. 2199-2210.
Huff, D.T., A.J. Weisman, and R. Jeraj, Interpretation and visualization techniques for deep learning models in medical imaging. Phys Med Biol, 2021. 66(4): p. 04tr01.
Krizhevsky, A., I. Sutskever, and G. Hinton, ImageNet classification with deep convolutional neural networks. Advances in neural information processing systems, 2012. 25(2).
Simonyan, K. and A. Zisserman, Very deep convolutional networks for large-scale image recognition. Computer Science, 2014.
He, K., et al., Deep residual learning for image recognition. IEEE, 2016.
Chollet, F., Xception: Deep learning with depthwise separable convolutions. IEEE, 2017.
Zhou, B., et al., Learning Deep features for discriminative localization. IEEE Computer Society, 2016.
Springenberg∗, J., et al., Striving for simplicity: the all convolutional net. eprint arxiv, 2014.
Litjens, G., et al., A survey on deep learning in medical image analysis. Med Image Anal, 2017. 42: p. 60-88.
Di, J., S. Rutherford, and C. Chu, Review of the cervical cancer burden and population-based cervical cancer screening in China. Asian Pac J Cancer Prev, 2015. 16(17): p. 7401-7.
Cao, L., et al., A novel attention-guided convolutional network for the detection of abnormal cervical cells in cervical cancer screening. Med Image Anal, 2021. 73: p. 102197.
Chandran, V., et al., Diagnosis of cervical cancer based on ensemble deep learning network using colposcopy images. Biomed Res Int, 2021. 2021: p. 5584004.
Wu, M., et al., Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci Rep, 2018. 38(6).
Adadi, A. and M. Berrada, Peeking inside the black-box: a survey on Explainable Artificial Intelligence (XAI). IEEE Access, 2018. 6: p. 52138-52160.
Selvaraju, R.R., et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. in Proceedings of the IEEE international conference on computer vision. 2017.
Aditya, C., et al., Grad-CAM++: improved visual explanations for deep convolutional networks. arXiv 2018. arXiv preprint arXiv:1710.11063.
Jahn, S.W., M. Plass, and F. Moinfar, Digital pathology: advantages, limitations and emerging perspectives. J Clin Med, 2020. 9(11).
Ying, X. and T.M. Monticello, Modern imaging technologies in toxicologic pathology: an overview. Toxicol Pathol, 2006. 34(7): p. 815-26.
Funding
This research was financially supported by the Chongming district Innovation and Entrepreneurship Project (granted by Mei Wang).
Author information
Authors and Affiliations
Contributions
LYX and CF contributed equally to this research. LYX, CF, and WM devised the study plan. LYX, CF, SJJ, and HYL helped with data acquisition. LYX, CF, and SJJ wrote the draft manuscript. WM and CF supervised the research.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The study protocol was approved by the institutional review board of Xinhua hospital (CMEC-2022-KT-15). Written informed consent was waived by the institutional review boards owing to the retrospective study design. However, verbal informed consent was obtained from all patients.
Consent for Publication
The authors affirm that verbal informed consent for publication was obtained from all patients.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Yx., Chen, F., Shi, Jj. et al. Convolutional Neural Networks for Classifying Cervical Cancer Types Using Histological Images. J Digit Imaging 36, 441–449 (2023). https://doi.org/10.1007/s10278-022-00722-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-022-00722-8