Abstract
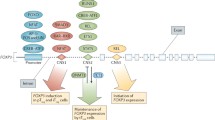
Forkhead box M1 (FoxM1), a proliferation specific transcriptional modulator, plays a principal role in many physiological and pathological processes. FoxM1-mediated oncogenic processes have been well addressed. However, functions of FoxM1 in immune cells are less summarized. The literatures about the expression of FoxM1 and its regulation on immune cells were searched on PubMed and Google Scholar. In this review, we provide an overview on the roles of FoxM1 in regulating functions of immune cells, including T cells, B cells, monocytes, macrophages, and dendritic cells, and discuss their contributions to diseases.


Similar content being viewed by others
Availability of data and material
Not applicable.
References
Wang J, Li W, Zhao Y, et al. Members of FOX family could be drug targets of cancers. Pharmacol Ther. 2018;181:183–96. https://doi.org/10.1016/j.pharmthera.2017.08.003.
Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143:4558–70. https://doi.org/10.1242/dev.112672.
Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genom. 2010;4:345–52. https://doi.org/10.1186/1479-7364-4-5-345.
Herman L, Todeschini AL, Veitia RA. Forkhead transcription factors in health and disease. Trends Genet. 2021;37:460–75. https://doi.org/10.1016/j.tig.2020.11.003.
Kalathil D, John S, Nair AS. FOXM1 and cancer: faulty cellular signaling derails homeostasis. Front Oncol. 2020;10:626836. https://doi.org/10.3389/fonc.2020.626836.
Sher G, Masoodi T, Patil K, et al. Dysregulated FOXM1 signaling in the regulation of cancer stem cells. Semin Cancer Biol. 2022. https://doi.org/10.1016/j.semcancer.2022.07.009.
Rajamanickam S, Panneerdoss S, Gorthi A, et al. Inhibition of FoxM1-mediated DNA repair by imipramine blue suppresses breast cancer growth and metastasis. Clin Cancer Res. 2016;22:3524–36. https://doi.org/10.1158/1078-0432.CCR-15-2535.
Wang Y, Wu M, Lei Z, et al. Dysregulation of miR-6868-5p/FOXM1 circuit contributes to colorectal cancer angiogenesis. J Exp Clin Cancer Res. 2018;37:292. https://doi.org/10.1186/s13046-018-0970-5.
Ghosh S, Singh R, Vanwinkle ZM, et al. Microbial metabolite restricts 5-fluorouracil-resistant colonic tumor progression by sensitizing drug transporters via regulation of FOXO3-FOXM1 axis. Theranostics. 2022;12:5574–95. https://doi.org/10.7150/thno.70754.
Ren X, Shah TA, Ustiyan V, et al. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol Cell Biol. 2013;33:371–86. https://doi.org/10.1128/MCB.00934-12.
Xue L, Chiang L, He B, Zhao YY, Winoto A. FoxM1, a forkhead transcription factor is a master cell cycle regulator for mouse mature T cells but not double positive thymocytes. PLoS ONE. 2010;5:e9229. https://doi.org/10.1371/journal.pone.0009229.
Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74–84. https://doi.org/10.1016/j.semcancer.2017.08.009.
Korver W, Roose J, Heinen K, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997;46:435–42. https://doi.org/10.1006/geno.1997.5065.
Gartel AL. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 2017;77:3135–9. https://doi.org/10.1158/0008-5472.CAN-16-3566.
Zhang YL, Ma Y, Zeng YQ, et al. A narrative review of research progress on FoxM1 in breast cancer carcinogenesis and therapeutics. Ann Transl Med. 2021;9:1704. https://doi.org/10.21037/atm-21-5271.
Li Y, Wu F, Tan Q, et al. The multifaceted roles of FOXM1 in pulmonary disease. Cell Commun Signal. 2019;17:35. https://doi.org/10.1186/s12964-019-0347-1.
Littler DR, Alvarez-Fernandez M, Stein A, et al. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010;38:4527–38. https://doi.org/10.1093/nar/gkq194.
Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol Chem. 2006;387:949–62. https://doi.org/10.1515/BC.2006.119.
Kalinichenko VV, Major ML, Wang X, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. https://doi.org/10.1101/gad.1200704.
Sanders DA, Gormally MV, Marsico G, et al. FOXM1 binds directly to non-consensus sequences in the human genome. Genome Biol. 2015;16:130. https://doi.org/10.1186/s13059-015-0696-z.
Kopanja D, Chand V, O’Brien EM, et al. Transcriptional repression by FoxM1 suppresses tumor differentiation and promotes metastasis of breast cancer. Cancer Res. 2022. https://doi.org/10.1158/0008-5472.CAN-22-0410.
Wang IC, Chen YJ, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. https://doi.org/10.1128/MCB.25.24.10875-10894.2005.
Liu C, Barger CJ, Karpf AR. FOXM1: a multifunctional oncoprotein and emerging therapeutic target in ovarian cancer. Cancers (Basel). 2021. https://doi.org/10.3390/cancers13123065.
Bella L, Zona S, Nestal de Moraes G, Lam EW. FOXM1: a key oncofoetal transcription factor in health and disease. Semin Cancer Biol. 2014;29:32–9. https://doi.org/10.1016/j.semcancer.2014.07.008.
Goldschneider I. Cyclical mobilization and gated importation of thymocyte progenitors in the adult mouse: evidence for a thymus-bone marrow feedback loop. Immunol Rev. 2006;209:58–75. https://doi.org/10.1111/j.0105-2896.2006.00354.x.
Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–76. https://doi.org/10.1146/annurev.immunol.21.120601.141107.
Takaba H, Takayanagi H. The mechanisms of T cell selection in the thymus. Trends Immunol. 2017;38:805–16. https://doi.org/10.1016/j.it.2017.07.010.
Enouz S, Carrie L, Merkler D, Bevan MJ, Zehn D. Autoreactive T cells bypass negative selection and respond to self-antigen stimulation during infection. J Exp Med. 2012;209:1769–79. https://doi.org/10.1084/jem.20120905.
Korver W, Roose J, Wilson A, Clevers H. The winged-helix transcription factor Trident is expressed in actively dividing lymphocytes. Immunobiology. 1997;198:157–61. https://doi.org/10.1016/S0171-2985(97)80036-8.
Starbeck-Miller GR, Xue HH, Harty JT. IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med. 2014;211:105–20. https://doi.org/10.1084/jem.20130901.
Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. https://doi.org/10.1016/j.bbcan.2006.08.006.
Laoukili J, Kooistra MR, Bras A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. https://doi.org/10.1038/ncb1217.
Leung TW, Lin SS, Tsang AC, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. https://doi.org/10.1016/s0014-5793(01)02915-5.
Wilkens AB, Fulton E, Pont MJ, et al. NOTCH1 signaling during CD4+ T-cell activation alters transcription factor networks and enhances antigen responsiveness. Blood. 2022. https://doi.org/10.1182/blood.2021015144.
Mata M, Gerken C, Nguyen P, et al. Inducible activation of MyD88 and CD40 in CAR T cells results in controllable and potent antitumor activity in preclinical solid tumor models. Cancer Discov. 2017;7:1306–19. https://doi.org/10.1158/2159-8290.CD-17-0263.
Prinzing B, Schreiner P, Bell M, et al. MyD88/CD40 signaling retains CAR T cells in a less differentiated state. JCI Insight. 2020. https://doi.org/10.1172/jci.insight.136093.
Kondo T, Ando M, Nagai N, et al. The NOTCH-FOXM1 axis plays a key role in mitochondrial biogenesis in the induction of human stem cell memory-like CAR-T cells. Cancer Res. 2020;80:471–83. https://doi.org/10.1158/0008-5472.CAN-19-1196.
Melchers F. Checkpoints that control B cell development. J Clin Investig. 2015;125:2203–10. https://doi.org/10.1172/JCI78083.
LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–80. https://doi.org/10.1182/blood-2008-02-078071.
Pieper K, Grimbacher B, Eibel H. B-cell biology and development. J Allergy Clin Immunol. 2013;131:959–71. https://doi.org/10.1016/j.jaci.2013.01.046.
Buchner M, Park E, Geng H, et al. Identification of FOXM1 as a therapeutic target in B-cell lineage acute lymphoblastic leukaemia. Nat Commun. 2015;6:6471. https://doi.org/10.1038/ncomms7471.
Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–71. https://doi.org/10.1038/nri3795.
Zhang M, Iwata S, Hajime M, et al. Methionine commits cells to differentiate into plasmablasts through epigenetic regulation of BTB and CNC homolog 2 by the methyltransferase EZH2. Arthritis Rheumatol. 2020;72:1143–53. https://doi.org/10.1002/art.41208.
Akita K, Yasaka K, Shirai T, et al. Interferon alpha enhances B cell activation associated with FOXM1 induction: potential novel therapeutic strategy for targeting the plasmablasts of systemic lupus erythematosus. Front Immunol. 2020;11:498703. https://doi.org/10.3389/fimmu.2020.498703.
Lefebvre C, Rajbhandari P, Alvarez MJ, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:377. https://doi.org/10.1038/msb.2010.31.
Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–23. https://doi.org/10.1038/ni.2060.
Ngo VN, Davis RE, Lamy L, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–10. https://doi.org/10.1038/nature04687.
Chen X, Muller GA, Quaas M, et al. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227–36. https://doi.org/10.1128/MCB.00881-12.
Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–89. https://doi.org/10.1101/gad.181933.111.
Zhao B, Barrera LA, Ersing I, et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–606. https://doi.org/10.1016/j.celrep.2014.07.037.
Wang IC, Ustiyan V, Zhang Y, et al. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D). Oncogene. 2014;33:5391–6. https://doi.org/10.1038/onc.2013.475.
Ganguly S, Kuravi S, Alleboina S, et al. Targeted therapy for EBV-associated B-cell neoplasms. Mol Cancer Res. 2019;17:839–44. https://doi.org/10.1158/1541-7786.MCR-18-0924.
Consolaro F, Basso G, Ghaem-Magami S, Lam EW, Viola G. FOXM1 is overexpressed in B-acute lymphoblastic leukemia (B-ALL) and its inhibition sensitizes B-ALL cells to chemotherapeutic drugs. Int J Oncol. 2015;47:1230–40. https://doi.org/10.3892/ijo.2015.3139.
Kuttikrishnan S, Prabhu KS, Khan AQ, et al. Thiostrepton inhibits growth and induces apoptosis by targeting FoxM1/SKP2/MTH1 axis in B-precursor acute lymphoblastic leukemia cells. Leuk Lymphoma. 2021;62:3170–80. https://doi.org/10.1080/10428194.2021.1957873.
Boer JM, den Boer ML. BCR-ABL1-like acute lymphoblastic leukaemia: from bench to bedside. Eur J Cancer. 2017;82:203–18. https://doi.org/10.1016/j.ejca.2017.06.012.
Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol. 2015;33:643–75. https://doi.org/10.1146/annurev-immunol-032414-112220.
Anderson NR, Minutolo NG, Gill S, Klichinsky M. Macrophage-based approaches for cancer immunotherapy. Cancer Res. 2021;81:1201–8. https://doi.org/10.1158/0008-5472.CAN-20-2990.
Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Mol Cancer. 2021;20:24. https://doi.org/10.1186/s12943-021-01313-x.
Atri C, Guerfali FZ, Laouini D. Role of human macrophage polarization in inflammation during infectious diseases. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19061801.
Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. https://doi.org/10.1016/j.cell.2010.03.014.
Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–62. https://doi.org/10.1016/j.immuni.2016.02.015.
Yang Y, Zhang B, Yang Y, Peng B, Ye R. FOXM1 accelerates wound healing in diabetic foot ulcer by inducing M2 macrophage polarization through a mechanism involving SEMA3C/NRP2/Hedgehog signaling. Diabetes Res Clin Pract. 2021;184:109121. https://doi.org/10.1016/j.diabres.2021.109121.
Goda C, Balli D, Black M, et al. Loss of FOXM1 in macrophages promotes pulmonary fibrosis by activating p38 MAPK signaling pathway. PLoS Genet. 2020;16:e1008692. https://doi.org/10.1371/journal.pgen.1008692.
Balli D, Ren X, Chou FS, et al. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene. 2012;31:3875–88. https://doi.org/10.1038/onc.2011.549.
Daemen S, Gainullina A, Kalugotla G, et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 2021;34:108626. https://doi.org/10.1016/j.celrep.2020.108626.
Rahman K, Vengrenyuk Y, Ramsey SA, et al. Inflammatory Ly6Chi monocytes and their conversion to M2 macrophages drive atherosclerosis regression. J Clin Investig. 2017;127:2904–15. https://doi.org/10.1172/JCI75005.
Ren X, Zhang Y, Snyder J, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30:5381–93. https://doi.org/10.1128/MCB.00876-10.
Sawaya AP, Stone RC, Brooks SR, et al. Deregulated immune cell recruitment orchestrated by FOXM1 impairs human diabetic wound healing. Nat Commun. 2020;11:4678. https://doi.org/10.1038/s41467-020-18276-0.
Hirani D, Alvira CM, Danopoulos S, et al. Macrophage-derived IL-6 trans-signalling as a novel target in the pathogenesis of bronchopulmonary dysplasia. Eur Respir J. 2022. https://doi.org/10.1183/13993003.02248-2020.
Xia H, Ren X, Bolte CS, et al. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am J Respir Cell Mol Biol. 2015;52:611–21. https://doi.org/10.1165/rcmb.2014-0091OC.
Kuznetsova T, Prange KHM, Glass CK, de Winther MPJ. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat Rev Cardiol. 2020;17:216–28. https://doi.org/10.1038/s41569-019-0265-3.
Szpak D, Izem L, Verbovetskiy D, et al. alphaMbeta2 is antiatherogenic in female but not male mice. J Immunol. 2018;200:2426–38. https://doi.org/10.4049/jimmunol.1700313.
Gage MC, Becares N, Louie R, et al. Disrupting LXRalpha phosphorylation promotes FoxM1 expression and modulates atherosclerosis by inducing macrophage proliferation. Proc Natl Acad Sci USA. 2018;115:E6556–65. https://doi.org/10.1073/pnas.1721245115.
Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. https://doi.org/10.1146/annurev.immunol.18.1.767.
Gardner A, de Mingo Pulido A, Ruffell B. Dendritic cells and their role in immunotherapy. Front Immunol. 2020;11:924. https://doi.org/10.3389/fimmu.2020.00924.
Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–92. https://doi.org/10.1146/annurev.immunol.021908.132557.
Wan H, Dupasquier M. Dendritic cells in vivo and in vitro. Cell Mol Immunol. 2005;2:28–35.
Worbs T, Hammerschmidt SI, Forster R. Dendritic cell migration in health and disease. Nat Rev Immunol. 2017;17:30–48. https://doi.org/10.1038/nri.2016.116.
Zhou Z, Chen H, Xie R, et al. Epigenetically modulated FOXM1 suppresses dendritic cell maturation in pancreatic cancer and colon cancer. Mol Oncol. 2019;13:873–93. https://doi.org/10.1002/1878-0261.12443.
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 82072927).
Author information
Authors and Affiliations
Contributions
JZ and PZ drafted the main body of this manuscript and drew the figures. XB, XW and XM modified the manuscript. PZ and XM took primary responsibility for this paper as the corresponding author. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors gave their consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, J., Bu, X., Wei, X. et al. The role of FoxM1 in immune cells. Clin Exp Med 23, 1973–1979 (2023). https://doi.org/10.1007/s10238-023-01037-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01037-w