Abstract
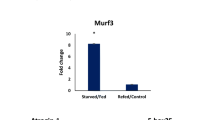
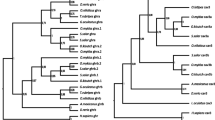
The selection of proteins destined for degradation by the ubiquitin–proteasome pathway is coordinated by E3 ubiquitin ligases (E3Ub). One group of E3Ubs is described as muscle-specific RING finger (MuRF) molecules. In mammals, these proteins are believed to be central to targetting of muscle proteins for degradation during physiological perturbations such as starvation and inflammatory responses. In fish, the diversity of MuRF sequences is unexplored as is the expression of their mRNAs. In this study, three MuRF1 cDNAs, denoted as MuRF1a, MuRF1b, and MuRF1c, and a single MuRF2 were identified and characterized in Atlantic salmon. The MuRF1 sequences are highly conserved and encode predicted proteins of 349, 350, and 353 amino acids, whereas MuRF2 encodes a longer protein of 462 amino acids. The evolutionary relationship of these sequences with other fish and mammalian molecules shows that MuRF1a and 1b may have arisen from a recent salmonid duplication. The mRNA of MuRFs was expressed in multiple tissues, with highest abundance in white muscle tissue followed by the heart. The expression of MuRFs was modulated after both starvation and immune challenge. Starvation increased expression of all MuRF mRNAs in white muscle, with the greatest increase found in MuRF1a. A proinflammatory stimulation increased expression of MuRF mRNA in muscle and other tissues indicating a role of these proteins in protein degradation during inflammation.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic Local Alignment Search Tool. J Mol Biol 215:403–410
Argiles JA, Lopez-Soriano FJ, Busquets S (2008) Apoptosis signalling is essential and precedes protein degradation in wasting skeletal muscle during catabolic conditions. Int J Biochem Cell Biol 40:1674–1678
Attaix D, Baracos VE (2010) MAFbx/atrogin-1 expression is a poor index of muscle proteolysis. Curr Opin Clin Nutr Metab Care 13:223–224
Attaix D, Combaret L, Pouch MN, Taillandier D (2001) Regulation of proteolysis. Curr Opin Clin Nutr Metab Care 4:45–49
Attaix D, Mosoni L, Dardevet D, Combaret L, Mirand PP, Grizard J (2005) Altered responses in skeletal muscle protein turnover during aging in anabolic and catabolic periods. Int J Biochem Cell Biol 37:1962–1973
Baumann H, Gauldie J (1994) The acute-phase response. Immunol Today 15:74–80
Bodine SC, Latres E, Baumhueter S, Lai VKM, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na EQ, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704–1708
Boonyarom O, Inui K (2006) Atrophy and hypertrophy of skeletal muscles: structural and functional aspects. Acta Physiol 188:77–89
Borden KLB, Freemont PS (1996) The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol 6:395–401
Bower NI, Johnston IA (2009) Selection of reference genes for expression studies with fish myogenic cell cultures. BMC Molecular Biology 10
Bower NI, Taylor RG, Johnston IA (2009) Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Frontiers in Zoology 6
Bower NI, de la Serrana DG, Johnston IA (2010) Characterisation and differential regulation of MAFbx/Atrogin-1 alpha and beta transcripts in skeletal muscle of Atlantic salmon (Salmo salar). Biochem Biophys Res Commun 396:265–271
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4
Cardozo T, Pagano M (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 5:739–751
Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S (2001) Identification of muscle specific RING finger proteins as potential regulators of the titin kinase domain. J Mol Biol 306:717–726
Chang W, Webster DR, Salam AA, Gruber D, Prasad A, Eiserich JP, Bulinski JC (2002) Alteration of the C-terminal amino acid of tubulin specifically inhibits myogenic differentiation. J Biol Chem 277:30690–30698
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Ciechanover A (1994) The ubiquitin–proteasome proteolytic pathway. Cell 79:13–21
Cleveland BM, Evenhuis JP (2010) Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): Expression across tissues in response to feed deprivation. Comp Biochem Physiol B Biochem Mol Biol 157:248–257
Cleveland BM, Weber GM, Blemings KP, Silverstein JT (2009) Insulin-like growth factor-I and genetic effects on indexes of protein degradation in response to feed deprivation in rainbow trout (Oncorhynchus mykiss). Am J Physiol Regul Integr Comp Physiol 297:R1332–R1342
Dinarello CA (1996) Biologic basis for interleukin-1 in disease. Blood 87:2095–2147
Dobly A, Martin SAM, Blaney SC, Houlihan DF (2004) Protein growth rate in rainbow trout (Oncorhynchus mykiss) is negatively correlated to liver 20S proteasome activity. Comp Biochem Physiol A Mol Integr Physiol 137:75–85
Fauconneau B, Gray C, Houlihan DF (1995) Assessment of individual protein-turnover in 3 muscle types of rainbow-trout. Comp Biochem Physiol B Biochem Mol Biol 111:45–51
Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, Richardson JA, Bassel-Duby R, Olson EN (2007) Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Investig 117:2486–2495
Fraser KPP, Rogers AD (2007) Protein metabolism in marine animals: the underlying mechanism of growth advances in marine biology. Vol 52, pp 267–362
Freemont PS (2000) Ubiquitination: RING for destruction? Curr Biol 10:R84–R87
Glass DJ (2003) Molecular mechanisms modulating muscle mass. Trends Mol Med 9:344–350
Goldberg AL (1995) Functions of the proteasome—the lysis at the end of the tunnel. Science 268:522–523
Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL (2001) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98:14440–14445
Higgins AD, Silverstein JT, Engles J, Wilson ME, Rexroad CE, Blemings KP (2005) Starvation induced alterations in hepatic lysine metabolism in different families of rainbow trout (Oncorhynchus mykiss). Fish Physiol Biochem 31:33–44
Houlihan DF, Carter CG, McCarthy ID (1995) Protein turnover in animals. In: Wright P, Walsh P (eds) Nitrogen metabolism and excretion. CRC Press, Boca Raton, pp 1–29
Jackson PK, Eldridge AG (2002) The SCF ubiquitin ligase: an extended look. Mol Cell 9:923–925
Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C (2004) Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA 101:18135–18140
Kornitzer D, Ciechanover A (2000) Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol 182:1–11
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lecker SH, Goldberg AL (2002) Slowing muscle atrophy: putting the brakes on protein breakdown. J Physiol Lond 545:729
Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18:39–51
Li HH, Kedar V, Zhang CL, McDonough H, Arya R, Wang DZ, Patterson C (2004) Atrogin-1/rnuscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Investig 114:1058–1071
Lyndon AR, Houlihan DF (1998) Gill protein turnover: costs of adaptation. Comp Biochem Physiol Part A Mol Integr Physiol 119:27–34
Martin SAM, Blaney S, Bowman AS, Houlihan DF (2002) Ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss): effect of food deprivation. Pflügers Arch Eur J Physiol 445:257–266
Martin SAM, Douglas A, Houlihan DF, Secombes CJ (2010) Starvation alters the liver transcriptome of the innate immune response in Atlantic salmon (Salmo salar). BMC Genomics 11:418
McElhinny AS, Perry CN, Witt CC, Labeit S, Gregorio CC (2004) Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J Cell Sci 117:3175–3188
Meroni G, Diez-Roux G (2005) TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. BioEssays 27:1147–1157
Nikawa T, Ishidoh K, Hirasaka K, Ishihara I, Ikemoto M, Kano M, Kominami E, Nonaka I, Ogawa T, Adams GR, Baldwin KM, Yasui N, Kishi K, Takeda S (2004) Skeletal muscle gene expression in space-flown rats. FASEB J 18:522–524
Olsvik PA, Lie KK, Jordal AEO, Nilsen TO, Hordvik I (2005) Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Molecular Biology 6
Peragon J, Barroso JB, Garcia-Salguero L, Aranda F, de la Higuera M, Lupianez JA (1999) Selective changes in the protein-turnover rates and nature of growth induced in trout liver by long-term starvation followed by re-feeding. Mol Cell Biochem 201:1–10
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Sacheck JM, Hyatt JPK, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL (2007) Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 21:140–155
Salem M, Kenney PB, Rexroad CE, Yao JB (2006) Molecular characterization of muscle atrophy and proteolysis associated with spawning in rainbow trout. Comp Biochem Physiol, D: Genomics Proteomics 1:227–237
Seiliez I, Panserat S, Skiba-Cassy S, Fricot A, Vachot C, Kaushik S, Tesseraud S (2008) Feeding status regulates the polyubiquitination step of the ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss) muscle. J Nutr 138:487–491
Spate U, Schulze PC (2004) Proinflammatory cytokines and skeletal muscle. Curr Opin Clin Nutr Metab Care 7:265–269
Spencer JA, Eliazer S, Ilaria RL, Richardson JA, Olson EN (2000) Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J Cell Biol 150:771–784
Tacchi L, Bickerdike R, Secombes CJ, Pooley NJ, Urquhart KL, Collet B, Martin SAM (2010) Ubiquitin E3 ligase atrogin-1 (Fbox-32) in Atlantic salmon (Salmo salar): sequence analysis, genomic structure and modulation of expression. Comp Biochem Physiol B Biochem Mol Biol 157(4):364–373
Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA (2005) Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem 280:2847–2856
Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68:1015–1068
Volff JN (2005) Genome evolution and biodiversity in teleost fish. Heredity 94:280–294
Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S (2008) Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J 27:350–360
Wright PA, Campbell A, Morgan RL, Rosenberger AG, Murray BW (2004) Dogmas and controversies in the handling of nitrogenous wastes: expression of arginase Type I and II genes in rainbow trout: influence of fasting on liver enzyme activity and mRNA levels in juveniles. J Exp Biol 207:2033–2042
Acknowledgments
The research was supported by an industrial studentship between the University of Aberdeen and BioMar Ltd (for L. Tacchi) and a Royal Society Grant (RG080331).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tacchi, L., Bickerdike, R., Secombes, C.J. et al. Muscle-Specific RING Finger (MuRF) cDNAs in Atlantic Salmon (Salmo salar) and Their Role as Regulators of Muscle Protein Degradation. Mar Biotechnol 14, 35–45 (2012). https://doi.org/10.1007/s10126-011-9385-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-011-9385-4