Abstract
Photo biomodulation (PBM) as a non-invasive and safe treatment has been demonstrated the anti-inflammatory potential in a variety of cell types, including stem cells. However, further investigations using different laser parameters combined with more accurate methods such as quantitative measurement of inflammatory gene expression at the mRNA level are still necessary. The aim of this study was to evaluate the effect of 532 nm green laser on cell proliferation as well as expression of inflammatory genes in human adipose-derived mesenchymal stem cells (hADMSCs) using RNA sequencing (RNA-seq) technique and confirmatory RT-PCR. hADMSCs were cultured in DMEM low glocuse medium with 10% fetal bovine serum until the fourth passage. Cultured cells were divided in two groups: control group (no laser irradiation) and laser group, irradiated with 532 nm laser at 44 m J/cm2 with an output power of 50 mW and a density of 6 mW/cm2, every other day, 7 s each time. The cell viability was assessed using MTT assay 24 h after each irradiation on days 3, 5, and 7 after cell seeding, followed by performing RNA-seq and RT-PCR. The MTT assay showed that PBM increased cell proliferation on day 5 after irradiation compared to day 3 and decreased on day 7 compared to day 5. In addition, gene expression analysis in hADMSCs using RNA-seq revealed down-regulation of inflammatory genes including CSF2, CXCL2, 3, 5, 6, 8, and CCL2, 7. These results indicate that 532 nm PBM with the parameters used in this study has a time-dependent effect on hADMSCs proliferation as well as anti-inflammatory potential.
Graphical abstract

Similar content being viewed by others
Introduction
Inflammation is a natural physiological immune response in the face of conditions such as infection, trauma, and disease. In pathological conditions, immune cells invoked at the site of infection or injury initiate a variety of activities, including vascularization, release of pro-inflammatory mediators, and the induction of phagocytosis [1, 2]. Chronic inflammation leads to various autoimmune and chronic immune-related diseases, such as type II diabetes, atherosclerosis, cancer, neurodegenerative diseases, rheumatoid arthritis (RA), and multiple sclerosis (MS) [3, 4] with high morbidity and mortality due to lack of effective immune-modulatory and anti-inflammatory treatment [5]. Chronic inflammation has an important role in the onset and progression of neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease [2]. In addition, numerous inflammatory genes play a critical role in the pathogenesis of inflammation-related tumors, which are involved in many cancer-associated functions and pathways [6]. Rheumatoid arthritis and osteoarthritis are two common forms of autoimmune and chronic diseases also associated with inflammatory genes [7]. Various pro-inflammatory mediators, such as cytokines (e.g., IL-1β, IL-6, IL-18, TNF-α, and IFN-γ) and chemokines (e.g., CCL2, CCL3, and CXCL3, 5, 6, 8), are involved in the onset of systemic immune responses [1, 2].
Adipose tissue-derived stem cells (ADSCs) have recently been considered as a possible treatment in chronic inflammation [3, 8]. In fact, ADSCs exert anti-inflammatory and immunosuppressive effects by inhibiting the production of pro-inflammatory cytokines by activated macrophages as well as inducing apoptotic cell death [8]. They have the potential to treat inflammatory, immune-mediated, and ischemic conditions through migration to the sites of inflammation and cell-cell interactions between ADSCs and lymphocytes or production of soluble growth factors. Nowadays, numerous stem cell-based strategies have provided a promising treatment against cancer that increases tumor targeting [9, 10]. ADSCs have been shown to modulate the microenvironment of the tumor to suppress cancer cells. They can release mediators such as exosomes which migrate to tumor sites, and thus deliver drugs and target tumor cells effectively [11].
Photobiomodulation (PBM), also known as low-level laser therapy (LLLT), is a non-invasive, painless, and safe treatment with minimum side effects [12]. PBM uses visible or invisible near-infrared (NIR) region of the spectrum, with wavelengths of 450–1200 nm (either LED or laser), and output power of 1–500 mW [13, 14] to modulate cellular responses without any thermal effect [15, 16]. The mechanism of PBM action occurs through the absorption of light by photoreceptors in the mitochondria, which modulates biochemical and photochemical reactions [17, 18].
Cytochrome-C-oxidase as complex IV of electron transport chain of the mitochondria is the major mitochondrial chromophore of PBM in the range of red to NIR light from 600 to about 900 nm [15, 16, 19]. Light absorbance leads to production of adenosine triphosphate (ATP) [17], reactive oxygen species (ROS), release of nitric oxide (NO) [20], increased membrane potential, and downstream cellular signaling via ATP, cAMP, ROS, Ca2+, and NO to influence gene transcription [15] and modulate various transcription factors [18]. It has been shown that PBM can improve cell survival, decrease apoptosis, reduce oxidative stress, suppress inflammation, and promote mitochondrial function [16, 21], as well as enhance cell proliferation and migration, and induce stem cell differentiation [22].
In addition to several studies which reported enhancing the proliferation and differentiation of stem cells [13], growing body of literature indicates that PBM can change the expression of various genes and proteins. Among them, many anti- and pro-inflammatory genes have regulated [23,24,25,26,27]. For example, phototherapy at 630 nm (32 and 64 J/cm2) and 465 nm (16, 32, 64 J/cm2) in human nucleus pulposus (NP) cells, under inflammatory conditions, significantly suppressed the production of IL-6 protein and inhibited the expression of IL-8 (CXCL8) compared with untreated control cells [28]. Another study displayed increased levels of IL-6, IL-8, and TNF-α mRNA in human outer root sheath cells (hORSCs) using 660 nm light (2.42 mW/cm2), with 1, 3, 5, or 10 J/cm2 of energy, while 830 nm NIR light at 1 J/cm2 on hORSCs increased IL-6 mRNA at 10 J/cm2 and IL-8 mRNA at 5 J/cm2 [29]. Also, LLLT with 660 nm GaAlAs laser (1, 2, and 3 J/cm2) significantly enhanced CCL2, CXCL10, and TNF-α, mRNA, and protein expression in human monocyte cell line THP-1 [30]. In another study, 660 nm laser irradiation on LPS-treated MSCs significantly decreased mRNA expression levels of inflammatory cytokines such as IL-1β, IL-6, and IL-8 and increased the expression and secretion of anti-inflammatory cytokines including IL-4 and IL-10 compared to control cells [31].
Additionally,WS1 human skin fibroblasts were examined in response to irradiation with a 660 nm diode laser (5 J/cm2, 11 mW/cm2) and it revealed up-regulation of several inflammatory cytokines and chemokines including CD40LG, CXCL11, CXCL2, IFNG, IL-10, IL-2, and IL-4 and down-regulation of CXCL1, CXCL5, and IL-1β [32]. Likewise, in a study involving patients with facial wrinkles showed that the mRNA level of IL-1 and TNF-α increased while IL-6 decreased by 633 nm LED laser [33].
GaInAlAs laser irradiation at 660 nm on L929 fibroblast cells showed down-regulation of IL-6 mRNA expression at 5 J/cm2 [34]. The mRNA level of TNF-α and IL-1β demonstrated a marked decrease using 810 nm (5 J/cm2) irradiation in RA synoviocytes. Also 25 J/cm2 had significant decrease in the intracellular levels of TNF-a, IL-1β, and IL-8 protein [35]. Accordingly, it suggested that the use of red and NIR LLLT as an effective treatment leads to down-regulation of pro-inflammatory cytokines and chemokine engaged in inflammatory response.
However, PBM with green light have been less studied. A study showed that laser irradiation at 532 nm (1.5 J/cm2) in cultured human skin fibroblasts increased the expression of type I and III procollagen, TIMP1 and TIMP2, Hsp70, and IL-6 whereas reduced MMP1 and MMP2 expression [36]. Moreover, PBM with 525 nm light inhibited the protein expression of inflammatory cytokine IL-6 in human nucleus pulposus cells at 16, 32, and 64 J/cm2 [28]. Although several previous studies have been concerned on the laser-induced gene expression changes in stem cells, there are few studies that have examined the effect of PBM at transcriptional expression level. Thus, the purpose of the present study was to explore the anti-inflammatory and cellular proliferation effects of green laser irradiation on hADMSCs.
Material and methods
Cell culture
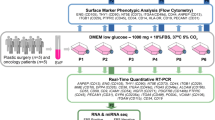
hADMSCs supplied from Stem Cell Technology Research Center were used in this experiment. The hADMSCs were cultured in a low-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; BioMEDIA) containing 10% fetal bovine serum (FBS; GibcoTM), 100 U/mL penicillin, and 100 µg/mL streptomycin. The cells were plated into 75 cm2 tissue culture flasks and incubated at 37 °C in a humidified atmosphere with 5% CO2, until reaching a confluence between 70 and 80%. The culture medium was changed every 3 days. Fourth passage culture of cells were washed with PBS (Bio IDEA), then trypsinized and were plated in 4-well plates for laser irradiation. The experiment involved two groups of cells: the control group was cultured in DMEM FBS 10% medium without laser irradiation and laser group was cultured in same condition with laser irradiation.
Laser irradiation
hADMSCs of laser group were plated in four 4-well plates at a density of 1 × 104 cell/well. A diode laser (Takfam Sazan Shafa, MODEL BS-310) was used in this study to generate a visible green laser beam with the wavelength of 532 nm, the power of 50 mW, and distance of 10 cm from the cells. The power density of the laser was 6 mW/cm2 and energy density was 44 mJ/cm2. The spot size of laser beam was 0.5 cm2, so we designed a beam expander for this experiment, to increase the irradiated surface area to 1.39 cm2 and cover the entire surface of each well of culture plate, uniformly. The total surface of the plate was irradiated three times in total, for 7 s, every other day. Nonirradiated control group were kept under the same conditions.
MTT assay
The MTT (3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide) assay is a colorimetric test performed for evaluating the cellular viability. The cells at a density of 1 × 104 cells/well from control and laser groups were seeded onto 4-well plates (4 well of each plate for each group), then were incubated for 24 h at 5% CO2 and 37 °C. After 24 h of incubation, MTT was dissolved in sterile PBS at a concentration of 5 mg/mL, added to the wells, and incubated for 4 h. Then the supernatant was removed and 100 µL DMSO added as a solubilization solution to dissolve the insoluble formazan product into a colored solution.
The absorbance of this solution was monitored by measuring at 570 nm by an ELISA reader. In this study, MTT assay was performed 24 h after each irradiation on days 3, 5, and 7 of cell seeding, with 4 replicates for each day.
RNA extraction and cDNA library construction
Total RNA was extracted from human adipose-derived mesenchymal stem cells in the control and laser groups, on day 7 of treatment, using Qiagen RNeasy Mini Kit, according to the manufacturer’s instructions (QIAGEN, Germany), then samples were quantified using a Nanodrop ND-1000 (Thermo Fisher Scientific) spectrophotometer. The integrity and concentration of isolated RNA were assessed using Agarose Gel Electrophoresis and Bioanalyzer 2100 (Agilent Technologies Inc., CA, USA) for evaluating the 28S and 18S ribosomal RNA bands (28S/18S ratio). The samples with RNA integrity number (RIN) of ≥8 were used for RNA library construction. RNA-seq library construction was performed using a Kapa Hyper Prep Kit (Kapa Biosystems) and subjected to 150 bp for sequencing.
RNA sequencing and data analysis
After library construction, RNA sequencing was performed on an Illumina HiSeq 4000 platform at Beijing Novogene Bioinformatics Technology Co., Ltd. (China). The quality control of sequencing reads was performed by FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to remove potential contaminations with adapters and low-quality and noisy data. After filtering the raw data, sequences were aligned to the Homo sapiens (human) genome, GRCh38 (hg38), using HISAT2 aligner v 2.1.0. Then FeatureCounts v1.5.0 was used to count the number of uniquely mapped read pairs with gene annotations. Differential gene expression values were determined using NOISeq v 2.22.0 based on an 80% probability threshold. Then differentially expressed genes (DEGs) with a log2 |FC| > = 1 were selected to compare the groups of study.
Real-time PCR
To confirm RNA-seq data on a selected number of genes, RT-PCR was performed on day 7 of treatment. Total RNA was extracted using the Qiagen RNeasy Mini Kit, according to the manufacturer’s instructions (QIAGEN, Germany) and 1 μg of total RNA was transcribed to complementary DNA (cDNA) using the Qiagen QuantiTect Reverse Transcription Kit. The quantitative real‐time polymerase chain reaction (qPCR) was performed on a Rotor‐Gene 6000 real‐time PCR machine (Corbett Research, Qiagen, Germany) under the following conditions: 95 °C for 90 s followed by 40 cycles, each cycle lasting 15 s at 95 °C and 30 s at 55 °C. Real-time PCR was performed in a 15 μL reaction mixture containing 7.5 μL of MasterMix SYBR Green PCR (Applied Biosystems), 0.75 μL of each primer (Table 1), 3 μL DNase- and RNase-free water, and 3 μL of cDNA from each sample. The primer sequences of the selected genes are listed in Table 1. GAPDH and β-actin were used as internal controls to normalize target gene expressions.
Statistical analysis
All experiments were performed with four replicates. Statistical analyses were performed using SPSS (IBM SPSS Statistics, USA) software. p-value < 0.05 was considered statistically significant.
Results
The effect of laser irradiation on cell viability
Following seeding of hADMSCs, the cells were irradiated by a 532 nm green laser at days D2, D4, and D6, as shown in Figs. 1 and 2.
Based on MTT assay data, the survival of MSCs after green laser irradiation shows a significant increase compared to the control group from day 3 to day 5 after treatment (p-value < 0.01) and a significant decrease compared to the control group from day 5 to day 7 after treatment (p-value < 0.01) (Fig. 3A).
A MTT assay shows a significant increase in optical density between the control and laser groups on day 3 and day 5 after treatment (*p-value < 0.05) (**p-value < 0.001). B Cell viability shows a significant increase)***p-value < 0.0005 ( from day 3 to 5 after irradiation and a significant decrease)**p-value < 0.001) from day 5 to day 7 after treatment. The values are expressed as means (± SEM; n = 4)
As shown in Fig. 3B, MTT assay shows that green light at 532 nm increased cell viability by 87%, 217%, and 103% (p-value < 0.01) on days 3, 5, and 7, respectively. However, cell viability showed a significant decrease on day 7 compared to day 5. The highest increase in survival compared to the control group was observed on the fifth day.
Differential gene expression analysis
To determine the DEGs in 532 nm laser irradiated cells, RNA-seq analysis was performed using Illumina HiSeq 4000 platform, 7 days after treatment. A total of 20.63 million and 20.75 million reads with a read length of 150 bp were aligned to Homo sapience reference genome (GRCh38) in the control and laser groups, respectively. 268 transcript IDs were identified as differentially expressed genes with probability threshold of ≥80%. Among them, 8 DEGs were related to inflammatory genes (Table 2) that all of them showed significant down-regulation. In addition, genes involved in cell cycle, proliferation, and apoptotic pathways such as CCND2 are down-regulated, while RIPK3, CCNA2, and CCNB2 are up-regulated.
Confirmation of DEGs by RT-PCR
In order to investigate the gene expression changes generated by RNA‐seq analysis, the expression of four DEGs was examined using qRT-PCR on day 7 in both groups. The selection criteria for these genes were based on the fold change, and previously LLLT studies. As shown in Tables 2 and 3, RNA-seq results were confirmed by qRT-PCR and these 4 genes were significantly decreased (p < 0.05) in line with our RNA‐seq data (Fig. 4; Table 3).
The expression levels for the candidate genes relative to β-actin as control (see Table 2). The values are expressed as means (± SEM; n = 3), (**p < 0.001); (*p < 0.05)
Discussion
Most PBM studies have shown that laser irradiation, especially in the red and NIR regions of the spectrum, can improve cell viability and proliferation, as documented in various cell types including fibroblasts, endothelial cells, skeletal cells, keratinocytes, myoblasts, and stem cells [37,38,39]. For example, 650 nm GaAlAs laser could increase the proliferation, differentiation, and secretion of the growth factors of the adipose-derived MSCs [40, 41] while PBM at 660 nm (5 mW, 6, 10, 12 J/cm2) could significantly increase the proliferation and viability of the bone marrow MSCs (BMSCs) [42]. In another study, low-level laser at 470 nm, 630 nm, and 660 nm promoted the proliferation and differentiation of BMSCs [43]. LLLT with 660 nm at 1–20 J/cm2 caused an increase in viability and protein concentration of human umbilical vein endothelial cells (HUVECs), while infrared laser (780 nm, 1–20 J/cm2) generally reduced cell viability [44]. In addition, the 660 and 810 nm laser at 3 J/cm2 can stimulate proliferation of hADSCs [45]. LED irradiation with 525, 660, and 830 nm wavelengths at 5 J/cm2 and 415, 525, 660, and 830 nm wavelengths at 10 J/ cm2 significantly enhanced outer root sheath cells (ORSCs) proliferation [29]. Our data, as shown by the MTT assay, indicated that 532 nm green light could increase ADSCs proliferation from day 3 to day 5 and decrease it from day 5 to day 7. This increase in cell proliferation on day 5 compared to day 3 can be explained by the increased expression of CCNA2 and CCNB2 according to RNA-seq and RT-PCR results. The protein encoded by CCNA2 (cyclin A2) acts as a regulator of the cell cycle. This protein binds and activates cyclin-dependent kinase 2 and enhances transition through G1/S and G2/M [46].
Additionally, the protein encoded by CCNB2, cyclin B2, is essential for the control of the cell cycle at the G2/M (mitosis) transition. Cyclin B2 is primarily associated with the Golgi region and also binds to transforming growth factor beta and thus cyclin B2/cdc2 may play a key role in transforming growth factor beta-mediated cell cycle control [47]. Therefore, it can be concluded that up-regulation of these two genes leads to increased proliferation from day 3 to 5 of treatment. As shown in Table 3, CCND2 showed a significant down-regulation according to the RNA-seq results (log2 FC = −1.75). The protein encoded by CCND2 (cyclinD2) functions as a regulatory subunit of the complex CDK4-cyclinD2 or CDK6-cyclinD2. The activity of cyclinD2 is required for cell cycle G1/S transition and interact with and be involved in the phosphorylation of tumor suppressor protein Rb [48]. This gene is also contributed in many biological processes such as positive regulation of cell population proliferation, positive regulation of G1/S mitotic cell cycle transduction, positive regulation of protein phosphorylation, and negative regulation of apoptotic process. Therefore, decreased expression of this gene on day 7 and after could explain why cell proliferation decreased compared to day 5 of treatment. We found green light at 532 nm also increased the expression of RIPK3 (receptor-interacting serine/threonine-protein kinase 3) gene (log2 FC = 1.85). The product of this gene is a member of the receptor-interacting protein (RIP) family of serine/threonine protein kinases, which activates necroptosis and apoptosis. It is a component of the tumor necrosis factor (TNF) receptor-I signaling complex, and can induce apoptosis and weakly activate the NF-κB transcription factor [47]. So we can conclude that increased expression of RIPK3 leads to decreasing in cell proliferation on day 7 comparing to day 5 of treatment.
Recent studies have shown that PBM plays an important role in regulating inflammatory gene expression and protein production in a variety of cells including stem cells [25,26,27,28,29,30,31,32,33,34,35]. But a few studies showed the effect of green light PBM as an inhibitory factor for inflammation on MSCs. Our results by RNA-seq and qRT-PCR suggested that 532 nm light could inhibit the expression of inflammatory genes like CSF2, CXCL2, 3, 5, 6, 8, and CCL2, 7.
CSF2, also known as granulocyte-macrophage colony-stimulating factor (GM-CSF), plays a role in promoting tissue inflammation and stimulates survival, production, differentiation, and function of hematopoietic precursor cells, including granulocytes and monocytes [49]. GM-CSF also has some effects on mature cells of the immune system, for example, enhancing neutrophil migration and causing an alteration of the receptors expressed on the cells surface [50]. Therefore, down-regulation of this gene showed anti-inflammatory effect of green laser therapy.
Our results showed down-regulation of 5 members of the CXC receptor ligand family. Among them, CXCL2 (C-X-C motif chemokine ligand 2) also known as macrophage inflammation protein 2 alpha (MIP-2α) [51] is a chemokine produced by activated monocytes and neutrophils that is involved in various immune and inflammatory processes. It is expressed at sites of inflammation and may suppress the proliferation of hematopoietic progenitor cells [48].
Another down-regulated gene, CXCL3 (C-X-C motif chemokine ligand 3), is an antimicrobial gene which encodes a protein that signals through the G-protein coupled receptor, CXC receptor 2. The encoded protein is involved in inflammation process and has chemotactic activity for neutrophils [52, 53]. Gene set enrichment analysis (GSEA) indicated that the overexpression of CXCL3 was closely associated with DNA repair, cell cycle process, cell apoptosis process, and the P53 regulation pathway [54]. This gene plays a multifaceted role in development and metastasis of various human cancers, such as uterine cervical cancer (UCC) [55], colon cancer (CC) [54], and prostate cancer [56]. In addition, CXCL3 and its receptor CXCR2 are overexpressed in prostate cancer cells, prostate epithelial cells, and prostate cancer tissues, which may play multiple roles in prostate cancer progression and metastasis [57]. Therefore, due to down-regulation of this gene by green laser light, green PBM can be considered as a potential treatment for these types of cancers.
CXCL5 is a member of the CXC subfamily of chemokines that has shown a significant down-regulation in both RNA-seq and RT-PCR data (log2 FC = −2.51). The protein encoded by this gene has been suggested to bind the G-protein coupled receptor chemokine (C-X-C motif) receptor 2 to recruit neutrophils, to promote angiogenesis, and to remodel connective tissues. This protein is thought to play a role in cancer cell proliferation, migration, and invasion [58]. In this regard, some in vitro and in vivo experiments revealed that CXCL5 is one of the important chemokines in tumor microenvironment and overexpression of CXCL5 is closely related to the survival time, recurrence, angiogenesis, and metastasis of cancers such as colorectal cancer (CRC) and gastric cancer [59,60,61,62]. CXCL5 also promotes hepatocellular carcinoma cells (HCC) proliferation and invasion, as well as intratumoral neutrophil infiltration [63]. Moreover, CXCL5 and CXCR2 expression levels increased in atherosclerotic coronary arteries plaque than in the normal coronary arteries [64].
Another down-regulated gene in our results is CXCL6 (C-X-C motif chemokine ligand 6) also known as granulocyte chemotactic protein 2 (GCP-2). It is a CXC chemokine expressed by epithelial cells of the airways, eyes, gastrointestinal tract, mammary glands, tonsils, macrophages, and mesenchymal cells, in particular during inflammation. It exerts chemotactic activity for neutrophil granulocytes and angiogenic properties. In addition, CXCL6 possess strong antibacterial activity against Gram-positive and Gram-negative bacteria, 90-fold higher than CXC chemokines CXCL5 and CXCL7 during localized infection [65]. CXCL6 has been shown to be a potent mediator in neo-angiogenesis [66] and can enhance tumor cell migration and invasion by accelerating MMP-9 activity, as well as tumor growth and metastasis [67]. In non-inflammatory conditions, CXCL6 in many cancers such as breast cancer [68], colorectal cancer [69, 70], osteosarcoma [70], non-small cell lung [71, 72], and endometrial cancer are highly expressed [67].
Finally, CXCL8 as the last member of the CXC receptor ligand family in our RNA-seq data had significant down-regulation (log2 FC = −4.26). The CXCL8 chemokine, also known as interleukin-8, is a major mediator of the inflammatory response and acts as a chemotactic agent for neutrophils, basophils, and T-cells, but not monocytes, to the site of infection. CXCL8 encode the interleukin-8 (IL-8) protein which is secreted by several cell types in response to an inflammatory stimulus including mononuclear macrophages, neutrophils, eosinophils, T lymphocytes, epithelial cells, and fibroblasts. This pro-inflammatory protein is thought to play a role in lung inflammation, coronary artery disease, and endothelial dysfunction. This protein promotes tumor cell proliferation, migration, invasion, angiogenesis, and metastasis in gastric and colon cancers [68, 73, 74]. Therefore, anti-CXCL8-targeted therapies for gastric and colon cancers can be an effective treatment [74].
CCL2 and CCL7 are two chemokines of the CC subfamily which showed down-regulation in RNA-seq data. CCL2 (C-C motif chemokine ligand 2) displays chemotactic activity for monocytes and basophils but not for neutrophils or eosinophils. It is involved in the pathogenesis of diseases such as psoriasis, rheumatoid arthritis [73], and atherosclerosis, which are characterized by monocyte recruitment and infiltration [75]. The overexpression of CCL2 is associated with severe acute respiratory syndrome coronavirus 2 infection [73] as well as increased tumor growth in breast [76, 77], ovarian [78], esophageal [79], gastric [80], renal cell [81], lung [82], colon [83], and papillary thyroid cancers [84,85,86]. Thus, CCL2 could be a potential therapeutic target for these cancers treatment.
CCL7 (CC motif chemokine ligand 7), also known as monocyte chemotactic protein 3 (MCP-3), is a CC chemokine that acts as a chemotactic factor and mediates the recruitment of various kinds of leukocytes, including monocytes, eosinophils, basophils, dendritic cells (DCs), NK cells, and activated T lymphocytes. CCL7 is involved in anti-inflammatory responses and tumorigenesis. CCL7 can promote tumor invasion and metastasis which leads to tumor progression [87]. CCL7 is up-regulated in metastatic renal cell carcinoma (RCC) [88], lung adenomas [87], and CRC [88, 89]. Increased expression of CCL7 also causes CRC metastasis in the liver [87, 90].
Conclusion
In conclusion, our results suggested that green light with a wavelength of 532 nm (44 mJ/cm2, 50 mW) has both enhancing and inhibitory effects on hADMSCs proliferation, in a timely dependent manner. We also found decreased expression of inflammatory cytokines and chemokines in laser irradiated cells compared with control which show anti-inflammatory potential of green laser light. In addition, considering the role of these inflammatory genes in various cancers and inflammatory diseases, it can be concluded that PBM with the parameters used in this study can be applied as a potential treatment in several stem cell therapy and regeneration approaches to reduce inflammation and enhance tissue repair as well as improving chronic inflammatory and cancers.
References
Fullerton JN, Gilroy DW (2016) Resolution of inflammation: a new therapeutic frontier. Nat Rev Drug Discovery 15(8):551–567. https://doi.org/10.1038/nrd.2016.39
Newcombe EA, Camats-Perna J, Silva ML, Valmas N, Huat TJ, and Medeiros R (2018) Inflammation: the link between comorbidities, genetics, and Alzheimer’s disease. J Neuroinflammation 15(1).https://doi.org/10.1186/s12974-018-1313-3
Petryk N, Shevchenko O (2020) Mesenchymal stem cells anti-inflammatory activity in rats: proinflammatory cytokines. J Inflamm Res 13:293–301. https://doi.org/10.2147/jir.s256932
Zhao Q, Ren H, Han Z (2016) Mesenchymal stem cells: immunomodulatory capability and clinical potential in immune diseases. J Cell Immunotherapy 2(1):3–20. https://doi.org/10.1016/j.jocit.2014.12.001
MacDonald ES, and Barrett JG (2020) The potential of mesenchymal stem cells to treat systemic inflammation in horses. Front Vet Sci 6. https://doi.org/10.3389/fvets.2019.00507
Jiang H, Dong L, Gong F, Gu Y, Zhang H, Fan D, Sun Z (2018) Inflammatory genes are novel prognostic biomarkers for colorectal cancer. Int J Mol Med. https://doi.org/10.3892/ijmm.2018.3631
Zhu N, Hou J, Wu Y, Li G, Liu J, Ma G, Song Y (2018) Identification of key genes in rheumatoid arthritis and osteoarthritis based on bioinformatics analysis. Medicine 97(22):e10997. https://doi.org/10.1097/md.0000000000010997
Kotani T, Masutani R, Suzuka T, Oda K, Makino S, and Ii M (2017) Anti-inflammatory and anti-fibrotic effects of intravenous adipose-derived stem cell transplantation in a mouse model of bleomycin-induced interstitial pneumonia. Sci Reports 7(1). https://doi.org/10.1038/s41598-017-15022-3
Chu D-T, Nguyen TT, Tien NLB, Tran D-K, Jeong J-H, Anh PG, … Dinh TC (2020) Recent progress of stem cell therapy in cancer treatment: molecular mechanisms and potential applications. Cells 9(3):563. https://doi.org/10.3390/cells9030563
Gomes JP, Assoni AF, Pelatti M, Coatti G, Okamoto OK, Zatz M (2017) Deepening a simple question: can MSCs be used to treat cancer? Anticancer Res 37:4747–4758. https://doi.org/10.21873/anticanres.11881
Aravindhan S, Ejam SS, Lafta MH, Markov A, Yumashev AV, Ahmadi M (2021) Mesenchymal stem cells and cancer therapy: insights into targeting the tumour vasculature. Cancer Cell Int 21:158. https://doi.org/10.1186/s12935-021-01836-9
Su C-T, Chen C-M, Chen C-C, Wu J-H (2020) Dose analysis of photobiomodulation therapy in stomatology. Evid-Based Complementary Altern Med 2020:1–12. https://doi.org/10.1155/2020/8145616
Chang S-Y, Carpena NT, Mun S, Jung JY, Chung P-S, Shim H, … Lee MY (2020) Enhanced inner-ear organoid formation from mouse embryonic stem cells by photobiomodulation. MolTher - Methods Clin Dev 17:556–567. https://doi.org/10.1016/j.omtm.2020.03.010
Austin E, Koo E, Merleev A, Torre D, Marusina A, Luxardi G, … Jagdeo J (2021) Transcriptome analysis of human dermal fibroblasts following red light phototherapy. Sci Reports, 11(1). https://doi.org/10.1038/s41598-021-86623-2
Liebert A, Bicknell B, Markman W, Kiat H (2020) A potential role for photobiomodulation therapy in disease treatment and prevention in the era of COVID-19. Aging Dis 11(6):1352–1362. https://doi.org/10.14336/AD.2020.0901
Bathini M, Raghushaker CR, Mahato KK (2020) The molecular mechanisms of action of photobiomodulation against neurodegenerative diseases: a systematic review. Cell Mol Neurobiol. https://doi.org/10.1007/s10571-020-01016-9
Martinelli A, Andreo L, Alves AN, Terena SML, Santos TC, Bussadori SK, … Mesquita-Ferrari RA (2020) Photobiomodulation modulates the expression of inflammatory cytokines during the compensatory hypertrophy process in skeletal muscle. Lasers Med Sci. https://doi.org/10.1007/s10103-020-03095-y
Ramezani F, Razmgir M, Tanha K, Nasirinezhad F, Neshasteriz A, Bahrami-Ahmadi A, … Janzadeh A (2020) Photobiomodulation for spinal cord injury: a systematic review and meta-analysis. Physiol Behavior, 112977. https://doi.org/10.1016/j.physbeh.2020.112977
Zhang D, Shen Q, Wu X, Xing D (2021) Photobiomodulation therapy ameliorates glutamatergic dysfunction in mice with chronic unpredictable mild stress-induced depression. Oxid Med Cell Longev. https://doi.org/10.1155/2021/6678276
Del Vecchio A, Tenore G, Luzi MC, Palaia G, Mohsen A, Pergolini D, Romeo U (2021) Laser photobiomodulation (PBM)—a possible new frontier for the treatment of oral cancer: a review of in vitro and in vivo studies. Healthcare 9(2):134. https://doi.org/10.3390/healthcare9020134
Heinig N, Schumann U, Calzia D, Panfoli I, Ader M, Schmidt MHH, Roehlecke C (2020) Photobiomodulation mediates neuroprotection against blue light induced retinal photoreceptor degeneration. Int J Mol Sci 21(7):2370. https://doi.org/10.3390/ijms21072370
Dompe C, Moncrieff L, Matys J, Grzech-Leśniak K, Kocherova I, Bryja A, Dyszkiewicz-Konwińska M (2020) Photobiomodulation—underlying mechanism and clinical applications. J Clin Med 9(6):1724. https://doi.org/10.3390/jcm9061724
Chen C-H, Tsai J-L, Wang Y-H, Lee C-L, Chen J-K, Huang M-H (2009) Low-level laser irradiation promotes cell proliferation and mRNA expression of type I collagen and decorin in porcine Achilles tendon fibroblasts in vitro. J Orthop Res 27(5):646–650. https://doi.org/10.1002/jor.20800
Usumez A, Cengiz B, Oztuzcu S, Demir T, Aras MH, Gutknecht N (2013) Effects of laser irradiation at different wavelengths (660, 810, 980, and 1,064 nm) on mucositis in an animal model of wound healing. Lasers Med Sci 29(6):1807–1813. https://doi.org/10.1007/s10103-013-1336-z
Yu W, Naim J, Lanzafame RJ (1997) Effects of photostimulation on wound healing in diabetic mice. Lasers Surg Med 20(1):56–63
Dadpay M, Sharifian Z, Bayat M, Bayat M, Dabbagh A (2012) Effects of pulsed infra-red low level-laser irradiation on open skin wound healing of healthy and streptozotocin-induced diabetic rats by biomechanical evaluation. J Photochem Photobiol B 111:1–8. https://doi.org/10.1016/j.jphotobiol.2012.03
Woodruff LD, Bounkeo JM, Brannon WM, Dawes KS, Barham CD, Waddell DL, Enwemeka CS (2004) The efficacy of laser therapy in wound repair: a meta-analysis of the literature. Photomed Laser Surg 22(3):241–247. https://doi.org/10.1089/1549541041438623
Hwang MH, Son HG, Lee JW, Yoo CM, Shin JH, Nam HG, … Choi H (2018) Phototherapy suppresses inflammation in human nucleus pulposus cells for intervertebral disc degeneration. Lasers Med Sci 33(5):1055–1064. https://doi.org/10.1007/s10103-018-2470-4
Kim JE, Woo YJ, Sohn KM, Jeong KH, Kang H (2017) Wnt/β-catenin and ERK pathway activation: a possible mechanism of photobiomodulation therapy with light-emitting diodes that regulate the proliferation of human outer root sheath cells. Lasers Surg Med 49(10):940–947. https://doi.org/10.1002/lsm.22736
Chen C-H, Wang C-Z, Wang Y-H, Liao W-T, Chen Y-J, Kuo C-H, Hung C-H (2014) Effects of low-level laser therapy on M1-related cytokine expression in monocytes via histone modification. Mediators Inflamm 2014:1–13. https://doi.org/10.1155/2014/625048
Yin K, Zhu R, Wang S, Zhao RC (2017) Low level laser (LLL) attenuate LPS-induced inflammatory responses in mesenchymal stem cells via the suppression of NF-κB signaling pathway in vitro. PLoS ONE 12(6):e0179175. https://doi.org/10.1371/journal.pone.0179175
Houreld NN, Ayuk SM, Abrahamse H (2014) Expression of genes in normal fibroblast cells (WS1) in response to irradiation at 660nm. J Photochem Photobiol B 130:146–152. https://doi.org/10.1016/j.jphotobiol.2013.11
Lee SY, Park K-H, Choi J-W, Kwon J-K, Lee DR, Shin MS, Park MY (2007) A prospective, randomized, placebo-controlled, double-blinded, and split-face clinical study on LED phototherapy for skin rejuvenation: clinical, profilometric, histologic, ultrastructural, and biochemical evaluations and comparison of three different treatment settings. J Photochem Photobiol B 88(1):51–67. https://doi.org/10.1016/j.jphotobiol.2007.04
Szezerbaty SKF, de Oliveira RF, Pires-Oliveira DAA, Soares CP, Sartori D, Poli-Frederico RC (2017) The effect of low-level laser therapy (660 nm) on the gene expression involved in tissue repair. Lasers Med Sci 33(2):315–321. https://doi.org/10.1007/s10103-017-2375-7
Yamaura M, Yao M, Yaroslavsky I, Cohen R, Smotrich M, Kochevar IE (2009) Low level light effects on inflammatory cytokine production by rheumatoid arthritis synoviocytes. Lasers Surg Med 41(4):282–290. https://doi.org/10.1002/lsm.20766
Dang Y, Ye X, Weng Y, Tong Z, Ren Q (2010) Effects of the 532-nm and 1,064-nm Q-switched Nd:YAG lasers on collagen turnover of cultured human skin fibroblasts: a comparative study. Lasers Med Sci 25(5):719–726. https://doi.org/10.1007/s10103-009-0657-4
Stein A, Benayahu D, Maltz L, Oron U (2005) Low-level laser irradiation promotes proliferation and differentiation of human osteoblasts in vitro. Photomed Laser Surg 23(2):161–166. https://doi.org/10.1089/pho.2005.23.161
Shefer G, Partridge TA, Heslop L, Gross JG, Oron U, Halevy O (2002) Low-energy laser irradiation promotes the survival and cell cycle entry of skeletal muscle satellite cells. J Cell Sci 115:1461–1469. https://doi.org/10.1242/jcs.115.7.1461
Jia Y-L, Guo Z-Y (2004) Effect of low-power He-Ne laser irradiation on rabbit articular chondrocytes in vitro. Lasers Surg Med 34(4):323–328. https://doi.org/10.1002/lsm.20017
Liao X, Li S-H, Xie G-H, Xie S, Xiao L-L, Song J-X, Liu H-W (2018) Preconditioning with low-level laser irradiation enhances the therapeutic potential of human adipose-derived stem cells in a mouse model of photoaged skin. Photochem Photobiol 94(4):780–790. https://doi.org/10.1111/php.12912
Ahrabi B, Rezaei Tavirani M, Khoramgah MS, Noroozian M, Darabi S, Khoshsirat S (2019) The effect of photobiomodulation therapy on the differentiation, proliferation, and migration of the mesenchymal stem cell: a review. J Lasers Med Sci 10(suppl 1):S96–S103. https://doi.org/10.15171/jlms.2019.s17
Cavalcanti MFXB, Maria DA, de Isla N, Leal-Junior ECP, Joensen J, Bjordal JM, … Frigo L (2015) Evaluation of the proliferative effects induced by low-level laser therapy in bone marrow stem cell culture. Photomed Laser Surg 33(12):610–616. https://doi.org/10.1089/pho.2014.3864
Ahn J-C, Rhee Y-H, Choi S-H, Kim DY, Chung P-S (2015) Low level light promotes the proliferation and differentiation of bone marrow derived mesenchymal stem cells. Mechanisms for Low-Light Therapy X. https://doi.org/10.1117/12.2078863
Terena SML, Mesquita-Ferrari RA, de Siqueira Araújo AM, Fernandes KPS, Fernandes MH (2020) Photobiomodulation alters the viability of HUVECs cells. Lasers Med Sci. https://doi.org/10.1007/s10103-020-03016-z
Wang Y, Huang Y-Y, Wang Y, Lyu P, and Hamblin MR (2017) Red (660 nm) or near-infrared (810 nm) photobiomodulation stimulates, while blue (415 nm), green (540 nm) light inhibits proliferation in human adipose-derived stem cells. Sci Reports 7(1). https://doi.org/10.1038/s41598-017-07525-w
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2008) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2014) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res
Sielska M, Przanowski P, Pasierbińska M, Wojnicki K, Poleszak K, Wojtas B, Kaminska B (2020) Tumour-derived CSF2/granulocyte macrophage colony stimulating factor controls myeloid cell accumulation and progression of gliomas. Br J Cancer 123(3):438–448. https://doi.org/10.1038/s41416-020-0862-2
Gasson JC (1991) Molecular physiology of granulocyte-macrophage colony-stimulating factor. Blood 77(6):1131–1145
Yang Y, Zhou X, Li Y, Chen A, Liang W, Liang G, Jin D (2019) CXCL2 attenuates osteoblasts differentiation by inhibiting ERK1/2 signaling pathway. J Cell Sci. https://doi.org/10.1242/jcs.230490
Ahuja SK, Murphy PM (1996) The CXC chemokines growth-regulated oncogene (GRO) α, GROβ, GROγ, neutrophil-activating peptide-2, and epithelial cell-derived neutrophil-activating peptide-78 are potent agonists for the type B, but not the type A, human interleukin-8 receptor. J Biol Chem 271(34):20545–20550. https://doi.org/10.1074/jbc.271.34.20545
Smith DF, Galkina E, Ley K, Huo Y (2005) GRO family chemokines are specialized for monocyte arrest from flow. Am J Physiol-Heart Circulatory Physiol 289(5):H1976–H1984. https://doi.org/10.1152/ajpheart.00153.2005
Ruan G, Gong Y, Liao X, Wang S, Huang W, Wang X, Gao F (2019) Diagnostic and prognostic values of C-X-C motif chemokine ligand 3 in patients with colon cancer. Oncol Rep. https://doi.org/10.3892/or.2019.7326
Qi Y, Li Y, Man X, Sui H, Zhao X, Zhang P, Wang W (2019) CXCL3 overexpression promotes the tumorigenic potential of uterine cervical cancer cells via the MAPK/ERK pathway. J Cell Physiol. https://doi.org/10.1002/jcp.29353
Xin H, Cao Y, Shao M, Zhang W, Zhang C, Wang J, Wang W (2018) Chemokine CXCL3 mediates prostate cancer cells proliferation, migration and gene expression changes in an autocrine/paracrine fashion. Int Urol Nephrol 50(5):861–868. https://doi.org/10.1007/s11255-018-1818-9
Gui S, Teng L, Wang S, Liu S, Lin Y-L, Zhao X, Wang W (2016) Overexpression of CXCL3 can enhance the oncogenic potential of prostate cancer. Int Urol Nephrol 48(5):701–709. https://doi.org/10.1007/s11255-016-1222-2
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2013) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res
Zhang W, Wang H, Sun M, Deng X, Wu X, Ma Y, Miao L (2020) CXCL5/CXCR2 axis in tumor microenvironment as potential diagnostic biomarker and therapeutic target. Cancer Commun 40(2–3):69–80. https://doi.org/10.1002/cac2.12010
Kawamura M, Toiyama Y, Tanaka K, Saigusa S, Okugawa Y, Hiro J, Kusunoki M (2012) CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur J Cancer 48(14):2244–2251. https://doi.org/10.1016/j.ejca.2011.11.032
Chen C, Xu Z-Q, Zong Y-P, Ou B-C, Shen X-H, Feng H, Lu A-G (2019) CXCL5 induces tumor angiogenesis via enhancing the expression of FOXD1 mediated by the AKT/NF-κB pathway in colorectal cancer. Cell Death Dis 10(3). https://doi.org/10.1038/s41419-019-1431-6
Park JY, Park KH, Bang S, Kim MH, Lee J-E, Gang J, Song SY (2007) CXCL5 overexpression is associated with late stage gastric cancer. J Cancer Res Clin Oncol 133(11):835–840. https://doi.org/10.1007/s00432-007-0225-x
Zhou S-L, Dai Z, Zhou Z-J, Wang X-Y, Yang G-H, Wang Z, Zhou J (2012) Overexpression of CXCL5 mediates neutrophil infiltration and indicates poor prognosis for hepatocellular carcinoma. Hepatology 56(6):2242–2254. https://doi.org/10.1002/hep.25907
Wang X-Z, Liu L-W, Du X-M, Gu R-X, Sun Z-J (2015) CXCL5 is associated with the increased risk of coronary artery disease. Coron Artery Dis 26(7):612–619. https://doi.org/10.1097/mca.0000000000000292
Linge HM, Collin M, Nordenfelt P, Morgelin M, Malmsten M, Egesten A (2008) The human CXC chemokine granulocyte chemotactic protein 2 (GCP-2)/CXCL6 possesses membrane-disrupting properties and is antibacterial. Antimicrob Agents Chemother 52(7):2599–2607. https://doi.org/10.1128/aac.00028-08
Keeley EC, Mehrad B, Strieter RM (2011) Chemokines as mediators of tumor angiogenesis and neovascularization. Exp Cell Res 317(5):685–690. https://doi.org/10.1016/j.yexcr.2010.10.020
Zhang H, Hou L, Li CM, Zhang WY (2013) The chemokine CXCL6 restricts human trophoblast cell migration and invasion by suppressing MMP-2 activity in the first trimester. Hum Reprod 28(9):2350–2362. https://doi.org/10.1093/humrep/det258
Lin C, He H, Liu H, Li R, Chen Y, Qi Y, Xu J (2019) Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut gutjnl–2018–316324. https://doi.org/10.1136/gutjnl-2018-316324
Xu L, Duda DG, di Tomaso E, Ancukiewicz M, Chung DC, Lauwers GY, Jain RK (2009) Direct evidence that bevacizumab, an anti-VEGF antibody, up-regulates SDF1, CXCR4, CXCL6, and neuropilin 1 in tumors from patients with rectal cancer. Can Res 69(20):7905–7910. https://doi.org/10.1158/0008-5472.can-09-2099
Yoshida T, Kobayashi T, Itoda M, Muto T, Miyaguchi K, Mogushi K, Tanaka H (2010) Clinical omics analysis of colorectal cancer incorporating copy number aberrations and gene expression data. Cancer Informatics 9:CIN.S3851. https://doi.org/10.4137/cin.s3851
Arenberg DA, Polverini PJ, Kunkel SL, Shanafelt A, Hesselgesser J, Horuk R, Strieter RM (1997) The role of CXC chemokines in the regulation of angiogenesis in non-small cell lung cancer. J Leukoc Biol 62(5):554–562. https://doi.org/10.1002/jlb.62.5.554
Lunghi L, Ferretti ME, Medici S, Biondi C, Vesce F (2007) Control of human trophoblast function. Reprod Biol Endocrinol 5(1):6. https://doi.org/10.1186/1477-7827-5-6
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2020) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res
Shen T, Yang Z, Cheng X, Xiao Y, Yu K, Cai X, Li Y (2017) CXCL8 induces epithelial-mesenchymal transition in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway. Oncol Rep 37(4):2095–2100. https://doi.org/10.3892/or.2017.5453
Li Y-S, Shyy Y-J, Wright JG, Valente AJ, Cornhill JF, Kolattukudy PE (1993) The expression of monocyte chemotactic protein (MCP-1) in human vascular endotheliumin vitro andin vivo. Mol Cell Biochem 126(1):61–68. https://doi.org/10.1007/bf01772208
Soria G, Ben-Baruch A (2008) The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett 267(2):271–285. https://doi.org/10.1016/j.canlet.2008.03.018
Soria G, Yaal-Hahoshen N, Azenshtein E, Shina S, Leider-Trejo L, Ryvo L, Ben-Baruch A (2008) Concomitant expression of the chemokines RANTES and MCP-1 in human breast cancer: a basis for tumor-promoting interactions. Cytokine 44(1):191–200. https://doi.org/10.1016/j.cyto.2008.08.002
Negus P et al (1995) The detection and localization of monocyte chemoattractant protein-1 (MCP-1) in human ovarian cancer. J Clin Invest 95(5):2391–2396. https://doi.org/10.1172/JCI117933
Ohta M, Kitadai Y, Tanaka S, Yoshihara M, Yasui W, Mukaida N, Chayama K (2002) Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human esophageal squamous cell carcinomas. Int J Cancer 102(3):220–224. https://doi.org/10.1002/ijc.10705
Ohta M, Kitadai Y, Tanaka S, Yoshihara M, Yasui W, Mukaida N, Chayama K (2003) Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human gastric carcinomas. Int J Oncol 22(4):773–778
Hemmerlein B, Johanns U, Kugler A, Reffelmann M, Radzun H-J (2001) Quantification and in situ localization of MCP-1 mRNA and its relation to the immune response of renal cell carcinoma. Cytokine 13(4):227–233. https://doi.org/10.1006/cyto.2000.0823
Niiya M, Niiya K, Kiguchi T, Shibakura M, Asaumi N, Shinagawa K, Tanimoto M (2003) Induction of TNF-α, uPA, IL-8 and MCP-1 by doxorubicin in human lung carcinoma cells. Cancer Chemother Pharmacol 52(5):391–398. https://doi.org/10.1007/s00280-003-0665-1
Huang S, Singh RK, Xie K, Gutman M, Berry KK, Bucana CD, Bar-Eli M (1994) Expression of theJE/MCP-1 gene suppresses metastatic potential in murine colon carcinoma cells. Cancer Immunol Immunother 39(4):231–238. https://doi.org/10.1007/bf01525986
Tanaka K, Kurebayashi J, Sohda M, Nomura T, Prabhakar U, Yan L, Sonoo H (2009) The expression of monocyte chemotactic protein-1 in papillary thyroid carcinoma is correlated with lymph node metastasis and tumor recurrence. Thyroid 19(1):21–25. https://doi.org/10.1089/thy.2008.0237
Hao Q, Vadgama JV, and Wang P (2020) CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal 18(1). https://doi.org/10.1186/s12964-020-00589-8
Arakaki R, Yamasaki T, Kanno T, Shibasaki N, Sakamoto H, Utsunomiya N, Kamba T (2016) CCL2 as a potential therapeutic target for clear cell renal cell carcinoma. Cancer Med 5(10):2920–2933. https://doi.org/10.1002/cam4.886
Yangyang L, Yadi C, Li L, Yudong W, Xiangyang X (2018) Crucial biological functions of CCL7 in cancer. PeerJ. https://doi.org/10.7717/peerj.4928
Wyler L, Napoli CU, Ingold B, Sulser T, Heikenwälder M, Schraml P, Moch H (2013) Brain metastasis in renal cancer patients: metastatic pattern, tumour-associated macrophages and chemokine/chemoreceptor expression. Br J Cancer 110(3):686–694. https://doi.org/10.1038/bjc.2013.755
Parikh N, Shuck RL, Gagea M, Shen L, Donehower LA (2017) Enhanced inflammation and attenuated tumor suppressor pathways are associated with oncogene-induced lung tumors in aged mice. Aging Cell 17(1):e12691. https://doi.org/10.1111/acel.12691
Cho YB, Lee WY, Choi S-J, Kim J, Hong HK, Kim S-H, Lee KU (2012) CC chemokine ligand 7 expression in liver metastasis of colorectal cancer. Oncol Reports 28(2):689–694. https://doi.org/10.3892/or.2012.1815
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tamimi, R., Mahmoodi, N.M., Samadikhah, H.R. et al. Anti-inflammatory effect of green photobiomodulation in human adipose-derived mesenchymal stem cells. Lasers Med Sci 37, 3693–3703 (2022). https://doi.org/10.1007/s10103-022-03654-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-022-03654-5