Abstract
To determine human bocavirus-1 (HBoV1) infection characteristics in young Australian children. Data were from the Observational Research in Childhood Infectious Diseases (ORChID) study, a Brisbane, Australia–based birth cohort of healthy, term, newborns followed prospectively for 2 years. Parents recorded daily symptoms, maintained an illness-burden diary, and collected weekly nasal swabs, which were tested for 17 respiratory viruses, including HBoV1, by real-time polymerase chain reaction (PCR) assays. Main outcomes measured were infection incidence, risk factors, symptoms, and healthcare use. One hundred fifty-eight children in the ORChID cohort provided 11,126 weekly swabs, of which 157 swabs were HBoV1 positive involving 107 incident episodes. Co-detections were observed in 65/157 (41.4%) HBoV1-positive swabs (or 41/107 [38.3%] infection episodes), principally with rhinovirus. Shedding duration was 1 week in 64.5% of episodes. The incidence of HBoV1 infections in the first 2 years of life was 0.58 episodes per child-year (95% confidence interval [CI] 0.47–0.71), including 0.38 episodes per child-year (95% CI 0.30–0.49) associated with respiratory symptoms. Recurrent episodes occurred in 18/87 (20.7%) children following their primary infection. In the first 2 years of life, incidence of HBoV1 episodes increased with age, during winter and with childcare attendance. Overall, 64.2% of HBoV1 episodes were symptomatic, with 26.4% having healthcare contact. Viral load estimates were higher when children were symptomatic than when asymptomatic (mean difference = 3.4; 95% CI 1.0–5.7 PCR cycle threshold units). After age 6 months, HBoV1 is detected frequently in the first 2 years of life, especially during winter. Symptoms are usually mild and associated with higher viral loads.
Similar content being viewed by others
Introduction
Human bocavirus-1 (HBoV1) is a DNA virus of the Parvoviridae family, which was first identified in 2005 in children diagnosed with an acute respiratory illness (ARI) [1]. Subsequent studies indicated > 90% of children are seropositive by 6 years of age [2]. Symptoms in young children associated with HBoV1 infection include fever, cough, rhinitis, and wheezing, leading to diagnoses of the common cold, acute otitis media, pneumonia, bronchiolitis, or acute asthma [1, 3, 4]. Nonetheless, its role in these illnesses remains controversial as experimental models are lacking and virus DNA shedding in some studies can persist for up to 6 to12 months following a primary HBoV1 infection [5,6,7]. Such prolonged shedding may compromise cross-sectional studies where HBoV1 is co-detected with other respiratory viruses in as many as 95% of cases [4, 8] and identified in 5 to 44% of asymptomatic children [5, 9,10,11]. Support for a pathogenic role comes from prospective studies identifying HBoV1 more frequently in children hospitalised with an ARI than age-matched asymptomatic controls [9, 10, 12]. There are also studies in symptomatic children where HBoV1 was the only virus identified, where increased HBoV1 viral load was associated with disease severity, and mRNA transcriptional activity or IgG seroconversion indicated acute infection [13,14,15]. Furthermore, HBoV1 was detected as the sole pathogen in the tracheal aspirates of a small number of mechanically ventilated children in the USA with severe lower respiratory infections [16], and in the lung aspirates of children in Sub-Saharan Africa with radiographic-confirmed pneumonia [17, 18].
HBoV1 is detected globally, and much of the epidemiological data are derived from cross-sectional hospital and laboratory-based studies [8,9,10, 12, 19,20,21,22,23,24,25,26]. However, to obtain a greater understanding of HBoV1 pathogenicity and its underlying epidemiology and disease burden, longitudinal community-based studies are needed to overcome the challenges posed by prolonged virus shedding. Where prospective, community-based HBoV1 studies exist [5,6,7, 11, 27, 28], they have had one or more limitations. These include being limited to ≤ 1-year duration [6, 11, 27, 28], to sampling principally during ARI episodes [5, 6, 27, 28], being confined to children attending childcare centres [5], with incomplete data capture of respiratory symptoms [7], possible sample contamination from other children [7], nasal swabs not collected routinely at the time of ARI episodes [11], and no reporting of other respiratory viruses [7]. Consequently, HBoV1 community-level epidemiology and disease burden in early life remains underexplored. The Australian Observational Research in Childhood Infectious Diseases (ORChID) birth cohort provided the opportunity to address many of these limitations by having parents record a daily symptom diary and collecting weekly nasal swabs from their newborn babies until their second birthday [29]. Previous reports from this community cohort identified that by age 2 years, 75% of participants had acquired HBoV1 on at least one occasion [30], although detections were not necessarily associated with ARI episodes [31]. Here, we examine in greater detail the characteristics of HBoV1 infections, including their epidemiology, risk factors, symptoms, and healthcare use in Australian children aged < 2 years from a subtropical, urban setting.
Materials and methods
Study design and participants
The ORChID study (ClinicalTrials.gov: NCT01304914) was a dynamic birth cohort study, which was conducted in the subtropical city of Brisbane, Australia, between September 2010 and October 2014. Healthy term, newborn babies without underlying congenital abnormalities or chronic disorders were progressively recruited from two metropolitan hospitals between September 2010 and October 2012 and followed until their second birthday [29,30,31]. Parents provided informed consent for their child’s enrolment post-birth. Parents collected weekly anterior nasal swabs from their child and completed a daily symptom diary. Children exited the study when parents stopped returning study material or on the child’s second birthday.
Child and family characteristics
Child and family characteristics were recorded at study entry, including information on the pregnancy and delivery. Breastfeeding and childcare attendance details were updated 3 monthly by telephone. Exclusive breastfeeding was the period from birth until introduction of solids or formula milk. Childcare was classified as formal, if outside the home by a regulated childcare service, or informal if non-regulated from friends or family.
Nasal swab collection
Parents were taught to collect bilateral anterior nasal swabs using a single plastic-shaft, rayon-budded swab, which was inserted into the anterior cavity of the nose and rubbed around the internal walls of the nares. After sampling from each nostril, the swab was then placed in a transport tube with a foam pad reservoir soaked in viral transport medium containing chloramphenicol and amphotericin B (Virocult MW950, Medical Wire & Equipment, Wiltshire, England). The initial nasal swab was collected shortly after birth and then on the same day each week, irrespective of symptoms. The swabs were returned to the laboratory in a sealable bag by surface mail at ambient temperatures. They took a median 3 days (interquartile range [IQR] 2–4 days) to reach the laboratory from the time of collection and where the swabs were processed and stored immediately at − 80 °C.
Laboratory testing
Nasal swabs were vortexed in 2 mL of phosphate buffered saline from which 200 uL was spiked with equine herpes virus-1, which acted as an extraction and inhibition control, before nucleic acid was extracted using the CAS1820 XtractorGene automated system (Qiagen, Australia). An additional internal control was endogenous retrovirus-3 (ERV3), which determined the presence of human DNA, thereby indicating successful extraction of nucleic acid from epithelial cells and overall higher sample quality [29, 32].
Swab samples were batch-tested for 17 respiratory viruses, which in addition to HBoV1 included rhinovirus, influenza viruses A and B, respiratory syncytial viruses A and B, parainfluenza viruses 1–3, human adenovirus, human metapneumovirus, human coronaviruses (OC43, HKU1, 229E, and NL63), and human polyomaviruses, WUPyV and KIPyV using previously validated real-time polymerase chain reaction (PCR) assays [29,30,31,32].
The HBoV1 assay used in the ORChID study for respiratory samples was a singleplex assay comprised of a single set of primers that amplified a 96-nucleotide sequence of the VP1 gene, and a TaqMan™ probe labelled with FAM and BHQ1 (Table 1). The 20-uL reaction mix comprised the following: 10 uL of SesiMix™ II probe mix (Bioline, Australia) and 2.0 uL of sample extract, 8 pmol of each primer, and 3.2 pmol of the TaqMan™ probe. The amplification was performed under the following thermo-cycling conditions: an activation step at 94 °C for 10 min, then 45 cycles of a denaturation step at 94 °C for 15 s, then annealing and elongation steps at 60 °C for 60 s. Acquisition of the PCR product signal occurred during the annealing and elongation step. All detections with cycle threshold (Ct) values ≤ 40 were considered positive for HBoV1 and other target viruses (Supplementary Fig. 1) [29,30,31,32].
PCR Ct values were also used as a semi-quantitative measure of viral loads as they are inversely proportional to the amount of target nucleic acid in the test sample. Thus lower Ct values align with higher viral load and a difference of 3.3 cycles corresponds to a tenfold difference in viral load [33]. As part of quality control, swab samples with ERV3 Ct values > 38 were defined as being of lower quality [31, 32].
A new HBoV1 episode was defined as either detecting HBoV1 for the first time, after at least two intervening negative swabs, or ≥ 30 day from the last HBoV1-positive swab.
Symptom and illness diaries
An ARI was defined according to parent-reported symptoms collected from a daily tick-box symptom diary. Symptoms were pre-defined, and parents were trained to recognise them. An upper respiratory illness (URI) was recorded if nasal congestion/discharge, dry cough, or physician-diagnosed otitis media were present. An acute lower respiratory illness (ALRI) was recorded if rattly breathing, wet (moist) cough, shortness of breath, wheeze, or physician-diagnosed pneumonia was present. When a URI episode progressed to an ALRI, the entire episode was categorised as an ARLI.
Parents completed an illness-burden diary documenting healthcare-seeking behaviour when the child had URI or ARLI symptoms. To minimise inconvenience, parents did not complete illness-burden diaries for either isolated nasal symptoms or dry cough as we assumed healthcare would not be sought. Completed symptom and illness-burden diaries were mailed each month to the research team. An ARI associated with HBoV1 was defined if symptoms were recorded within 7 days of a new HBoV1 detection [30, 31]. Emergency department and hospital records were reviewed at study completion.
Data analysis
Incidence rates of single new HBoV1 episodes, and associations between pre-defined risk factors and single new HBoV1 episodes, were calculated using mixed-effects Poisson regression models, with child included as a random effect and models adjusted for exposure-time.
Lower-quality swabs were excluded from incidence calculations to avoid underestimating incidence rates. The association between HBoV1-positive swabs and other respiratory virus detections was investigated using log-binomial regression. All multivariable models were adjusted for age, season of detection, presence of older children in the household, and childcare attendance. The association between symptomatic episodes and shedding duration or viral load was performed using linear regression. All analyses were conducted using Stata statistical software v16 (StataCorp, College Station, TX, USA).
Results
One-hundred fifty-eight children returned 11,126 swabs, and 154 provided 87,547 symptom diary-days of observation (78% of expected observation-days). This included 10,811 swabs (66% expected) matched to 82,036 diary-days from 154 children and 8101 higher-quality swabs from 157 children (Fig. 1). Participants were predominantly first-born children with tertiary-educated parents (Table 2).
There were 157 HBoV1-positive swabs from 87 children, of which 107 were incident episodes (Supplementary Table 1). Most (64.5%) shedding events were for 1 week. At least one other virus was co-detected in 65/157 (41.4%) HBoV1-positive swabs (or 41/107 [38.3%] infection episodes), and this was most frequently rhinovirus. However, after adjusting for potentially confounding variables, HBoV1 was not associated with co-detection with any other specific virus (Supplementary Table 2).
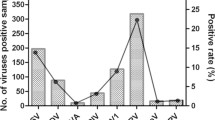
The overall incidence of HBoV1 in the first 2 years of life was 0.58 episodes per child-year (95% confidence interval [CI] 0.47–0.71). The incidence in the first and second years was 0.47 (95% CI 0.34–0.64) and 0.71 (95% CI 0.53–0.94) episodes per child-year, respectively. The overall incidence rate in the first 2 years of life for symptomatic HBoV1 infections was 0.38 (95% CI 0.30–0.49) episodes per child-year and 0.32 (95% CI 0.23–0.47) and 0.45 (95% CI 0.32–0.64) episodes per child-year in the first and second years of life, respectively. HBoV1 incidence increased with age during the first 2 years of life (Table 2), and episodes were more likely to occur in winter than summer seasons (adjusted incidence rate ratio (aIRR) = 3.0; 95% CI 1.6–5.8) (Table 2, Supplementary Fig. 2) and when the child attended childcare (aIRR = 2.2; 95% CI 1.3–3.8) (Table 2).
There were 106 incident episodes linked to symptom diaries, and 68 (64.2%) were associated with an ARI (Table 3). There were 51 (75.0%) URIs, of which 22 consisted of either dry cough or runny nose alone. Healthcare was sought for 28 (26.4%) episodes, consisting of 23 (21.7%) consulting a family physician only and three (2.8%) presenting to an emergency department (Table 3). There were no hospital admissions. Antibiotics were prescribed for 16 (23.5%) children seeking healthcare, one of whom had otitis media, while for the remainder the indication for antibiotics was not recorded. Healthcare utilisation rates were non-significantly higher for HBoV1 sole detections than when another virus was co-detected in the same episode (Supplementary Table 3). Symptomatic children had significantly lower Ct values (indicating higher viral loads) than asymptomatic children (mean [standard deviation] trough Ct values of 29.0 [6.1] versus 32.4 [5.3] respectively; a mean decrease of 3.4 (95%CI 1.0–5.7) Ct units) (Supplementary Table 4). In contrast, shedding duration was not associated with symptom status (mean difference − 0.1; 95% CI: − 0.5 to 0.3) weeks.
While 69/87 (79.3%) children had a single identified HBoV1 infection episode, 16 children had two discrete detection episodes, and a further two each had three episodes detected (Supplementary Fig. 3). The median time between episodes was 137 (IQR 66–183) days, which was well away from the 30-day threshold for defining a new detection episode. The shedding duration of HBoV1 DNA was longer following a primary infection episode than after subsequent detection episodes (mean [standard deviation] 1.7 [1.1] weeks versus 1.1 [0.2] weeks respectively, a mean difference of 0.6 [95%CI 0.4–0.8] weeks). In contrast, co-detection rates with other respiratory viruses when HBoV1 was first detected following primary infections (33/87; 37.9%) and subsequent episodes (8/20; 40.0%) were similar. In the 106 HBoV1 episodes linked to symptom diaries, 56/86 (65.1%) first episodes were associated with ARI symptoms, compared with 11/20 (55.0%) subsequent detection episodes (odds ratio = 0.76; 95% CI 0.29–2.20).
Discussion
HBoV1 was detected frequently in ORChID cohort participants during their first 2 years of life. Of the 17 respiratory viruses tested, it was the fourth most common virus after rhinovirus, human polyomaviruses WU/KI, and seasonal coronaviruses. Co-detections occurred in 38.3% of episodes, which is lower than the 52 to 95% reported for hospital and laboratory-based studies [8, 21, 23, 26, 34], but similar to the 47% co-detection rate reported in a Danish community-based cohort study [11]. It is possible that higher co-detection rates in cross-sectional studies are either from sicker patients, where the second virus is a recognised pathogen, or persistent HBoV1 DNA shedding from a prior infection.
HBoV1 DNA shedding in our study was ≤ 4 weeks in 106/107 (99%) episodes. Reasons for the shorter shedding duration observed in our cohort are uncertain, but could include differences in sample collection and quality, transport to the laboratory, and in-house assay performance affecting sensitivity for detecting low HBoV1 DNA levels in those with persistent shedding [29, 32, 35]. Indeed, like other DNA viruses, HBoV1 can establish latent infection within cells of the upper airways [1]. Therefore, a possible explanation for prolonged virus DNA shedding following primary infection is release of latent HBoV1 DNA from cells disrupted by inflammatory responses to other respiratory pathogens. In contrast, active infection is characterised by mRNA transcriptional activity for periods of no more than 2 weeks [15, 36].
In the current study, recurrences or new detection episodes were identified in 20.7% of children following their primary infection. Other longitudinal studies have reported recurrences in as many as 38% of young children [6, 7, 37], although sequencing revealed most were not from new HBoV1 variants [6, 7]. The Danish community-based birth cohort, which collected monthly nasal swabs from children in the first year of life, reported 68% of 50 HBoV1-infected children with ARI symptoms shed virus DNA for < 1 month, and for no longer than 4 months in the remaining infected participants [11]. However, in a US birth cohort where children provided weekly saliva samples from birth until 18 months of age, shedding events ≤ 30 days occurred in just 36% of 66 primary HBoV1 infections, with some children shedding the virus DNA for > 1 year [7]. Recurrent episodes occurred in 38% of children 21 to 420 days (median 48 days) following cessation of their primary episode, in which only 25% had a new variant detected [7]. Similar results were found in another prospective study of healthy children aged 6 months to 3 years who over a 1-year period had nasal swab specimens collected during an ARI or otitis media episode. [6] Shedding HBoV1 patterns were determined in 46 children. Overall, 28% cleared the virus DNA within 30 days, 46% had persistent or intermittent detection of the same strain for up to 265 days, 22% had no recurrences after their single positive episode, and just 4% had new HBoV1 subtypes detected in subsequent episodes [6]. Release of latent HBoV1 DNA from epithelial cells might also explain some of these recurrent episodes and the low detection rates of new HBoV1 variants on such occasions. Nevertheless, our study did not find evidence for increased virus co-detections associated with subsequent new HBoV1 detection episodes.
Consistent with seroprevalence and birth cohort studies [7, 11, 37, 38], HBoV1 was detected rarely (< 5%) in ORChID participants during their first 6 months of life, but detections increased rapidly thereafter, such that by age 2 years 75% had experienced at least one HBoV1 infection [30]. With the notable exception of rhinovirus, this temporal pattern of respiratory virus infections is observed in other birth cohort studies [38, 39]. Possible explanations for these observations include protection provided by transplacental maternal antibodies, breastfeeding, and young infants having less exposure than older children to others outside the household, including attending childcare [40, 41].
The incidence of 0.58 episodes per child-year was less than reported by a similar birth cohort study (0.73 episodes per child-year) that also collected weekly specimens [7]. However, most children in this cohort had commenced childcare by 6 months of age [7], which we found to be an independent risk factor for HBoV1. Indeed, across several birth cohort studies, childcare attendance has been found to be a consistent risk factor for ARIs and healthcare utilisation in the preschool years [38, 41,42,43,44].
Despite cross-sectional epidemiological studies and a recent longitudinal birth cohort study not finding evidence of HBoV seasonality after controlling for sampling frequency [4, 38], our 4-year study identified annual winter peaks, supporting comparable observations in other birth cohorts [7, 11]. The reasons for these conflicting observations are likely to be complex as seasonality and virus transmission can be determined by several factors, including human behaviour, the impact of temperature and humidity upon virus stability within the environment, as well as meteorological factors affecting host airway defences, and finally virus-virus interference [45, 46]. Of these, meteorological conditions and human behaviour are likely to be the main factors driving differences in seasonality [47].
In ORChID participants, 64.2% of those with new HBoV1 detections experienced ARI symptoms, 75.0% of whom had a mild URI. Nevertheless, 41.2% of these children had healthcare contact, 23.5% received antibiotics, but none was admitted to hospital. These proportions are similar to those observed with other common viruses detected in ORChID participants: parainfluenza (59%, 60%, 54%, 30%, 2%), seasonal coronaviruses (50%, 63%, 48%, 18%, 1%), and respiratory syncytial virus (73%, 43%, 57%, 32%, 3%) for ARI symptoms, URI symptoms, healthcare contact, antibiotic treatment, and hospitalisation, respectively [48,49,50]. These results for single HBoV1 detection episodes were not significantly different from episodes where other viruses were present. As noted previously in support of HBoV1 as a genuine respiratory pathogen [13], children with HBoV1 detections and ARI symptoms had significantly higher (approximately tenfold) viral loads (lower HBoV1 Ct values) than those without symptoms, although this did not extend to prolonging shedding duration.
The ORChID study, as an unselected community-based cohort, has several strengths from its high-density sampling over a 4-year period, optimising detection rates, seasonal effects, and virus shedding kinetics. However, it does have limitations. First, despite parents being trained to recognise symptoms, some may have been missed or misclassified. While doctor-diagnosed otitis media and pneumonia were recorded in the illness-burden diary, the indication for prescribing antibiotics was not captured systematically. Second, although we and others have shown that with sensitive PCR assays, parent-collected nasal swabs mailed into the laboratory have at least similar virus detection rates to those collected by healthcare workers [35, 51, 52], suboptimal swabbing techniques may have still missed virus detections [32]. We therefore removed 3025 (27%) lower-quality swabs from incidence rate calculations to avoid underestimation of rates due to misclassification bias. Third, to maintain cohort retention, we did not collect blood samples, and neither did we sequence HBoV1 isolates or seek evidence of HBoV1 mRNA transcriptional activity [15], to differentiate between new infection events and reactivation of latent virus following the primary infection. Finally, as is common with studies of this nature, participants came from small and advantaged households, and results may not generalise to other populations or settings.
In conclusion, HBoV1 was detected commonly in the ORChID cohort in the first 2 years of life and associated with increasing age, winter, and childcare attendance. Shedding duration was ≤ 4 weeks and co-detections occurred in 38.3% of infection episodes. Almost two-thirds of infections were symptomatic, and 41.2% of symptomatic episodes led to healthcare visits, but all were managed in the community. Higher viral loads (lower Ct values) were associated with ARI symptoms. These findings add further evidence for HBoV1 being a genuine respiratory pathogen.
Abbreviations
- ALRI:
-
Acute lower respiratory illness
- ARI:
-
Acute respiratory illness
- aIRR:
-
Adjusted incidence rate ratio
- CI:
-
Confidence interval
- Ct:
-
Cycle threshold
- ERV-3:
-
Endogenous retrovirus-3
- HBoV1:
-
Human bocavirus-1
- IQR:
-
Interquartile range
- ORChID:
-
Observational Research in Childhood Infectious Diseases
- PCR:
-
Polymerase chain reaction
- URI:
-
Upper respiratory illness
References
Jartti T, Hedman K, Jartti L, Ruuskanen O, Allander T, Söderlund-Venermo M (2012) Human bocavirus – the first 5 years. Rev Med Virol 22(1):46–64. https://doi.org/10.1002/rmv.720
Broccolo F, Falcone V, Esposito S, Toniolo A (2015) Human bocaviruses: possible etiologic role in respiratory infection. J Clin Virol 72:75–81. https://doi.org/10.1016/j.jcv.2015.09.008
Qiu J, Söderlund-Venermo M, Young NS (2017) Human parvoviruses. Clin Microbiol Rev 30(1):43–113. https://doi.org/10.1128/CMR.00040-16
Christensen A, Kesti O, Elenius V, Eskola AL, Døllner H, Altunbulakli C et al (2019) Human bocaviruses and paediatric infections. Lancet Child Adolesc Heal 3(6):418–426. https://doi.org/10.1016/S2352-4642(19)30057-4
Martin ET, Fairchok MP, Kuypers J, Magaret A, Zerr DM, Wald A et al (2010) Frequent and prolonged shedding of bocavirus in young children attending daycare. J Infect Dis 201(11):1625–1632. https://doi.org/10.1086/652405
Wagner JC, Pyles RB, Miller AL, Nokso-Koivisto J, Loeffelholz MJ, Chonmaitree T (2016) Determining persistence of bocavirus DNA in the respiratory tract of children by pyrosequencing. Pediatr Infect Dis J 35(5):471–476. https://doi.org/10.1097/INF.0000000000001058
Martin ET, Kuypers J, McRoberts JP, Englund JA, Zerr DM (2015) Human bocavirus 1 primary infection and shedding in infants. J Infect Dis 212(4):516–524. https://doi.org/10.1093/infdis/jiv044
do Amaral de Leon C, Amantea SL, Pilger DA, Cantarelli V (2013) Clinical and epidemiological profile of lower respiratory tract infections associated with human bocavirus. Pediatr Pulmonol 48(11):1112–1118. https://doi.org/10.1002/ppul.22732
García-García ML, Calvo C, Pozo F, Pérez-Breña P, Quevedo S, Bracamonte T et al (2008) Human bocavirus detection in nasopharyngeal aspirates of children without clinical symptoms of respiratory infection. Pediatr Infect Dis J 27(4):358–360. https://doi.org/10.1097/INF.0b013e3181626d2a
Rhedin S, Lindstrand A, Hjelmgren A, Ryd-Rinder M, Öhrmalm L, Tolfvenstam T et al (2015) Respiratory viruses associated with community-acquired pneumonia in children: matched case–control study. Thorax 70(9):847–853. https://doi.org/10.1136/thoraxjnl-2015-206933
Von Linstow M-L, Hogh M, Hogh B (2008) Clinical and epidemiologic characteristics of human bocavirus in Danish infants. Results from a prospective birth cohort study. Pediatr Infect Dis J 27(10):897–902. https://doi.org/10.1097/INF.0b013e3181757b16
Brieu N, Guyon G, Rodière M, Segondy M, Foulongne V (2008) Human bocavirus infection in children with respiratory tract disease. Pediatr Infect Dis J 27(11):969–973. https://doi.org/10.1097/INF.0b013e31817acfaa
Zhao B, Yu X, Wang C, Teng Z, Wang C, Shen J et al (2013) High human bocavirus load is associated with disease severity in children under five years of age. PLoS One 8(4):e62318. https://doi.org/10.1371/journal.pone.0062318
Nascimento-Carvalho AC, Vilas-Boas A-L, Fontoura M-SH, Xu M, Vuorinen T, Söderlund-Venermo M et al (2018) Serologically diagnosed acute human bocavirus 1 infection in childhood community-acquired pneumonia. Pediatr Pulmonol 53(1):88–94. https://doi.org/10.1002/ppul.23891
Xu M, Arku B, Jartti T, Koskinen J, Peltola V, Hedman K et al (2017) Comparative diagnosis of human bocavirus 1 respiratory infection with messenger RNA reverse-transcription polymerase chain reaction (PCR), DNA quantitative PCR, and serology. J Infect Dis 215(10):1551–1557. https://doi.org/10.1093/infdis/jix169
Tsitsiklis A, Osborne CM, Kamm J, Williamson K, Kalantar K, Dudas G et al (2022) Lower respiratory tract infections in children requiring mechanical ventilation: a multicentre prospective surveillance study incorporating airway metagenomics. Lancet Microbe 3(4):e284–e293. https://doi.org/10.1016/S2666-5247(21)00304-9
Carrol ED, Mankhambo LA, Guiver M, Banda DL, Denis B, Dove W et al (2011) PCR improves diagnostic yield from lung aspiration in Malawian children with radiologically confirmed pneumonia. PLoS One 6(6):e21042. https://doi.org/10.1371/journal.pone.0021042
Howie SRC, Morris GAJ, Tokarz R, Ebruke BE, Machuka EM, Ideh RC et al (2014) Etiology of severe childhood pneumonia in The Gambia, West Africa, determined by conventional and molecular microbiological analysis of lung and pleural aspirate samples. Clin Infect Dis 59(5):682–685. https://doi.org/10.1093/cid/ciu384
Ziemele I, Xu M, Vilmane A, Rasa-Dzelzkalēja S, Hedman K, Söderlund-Venermo M et al (2019) Serodiagnosis of human bocavirus 1 infection among hospitalised children with lower respiratory tract infection in Latvia. Proc Latv Acad Sci Sect B Nat Exact, Appl Sci 73(4):288–295. https://doi.org/10.2478/prolas-2019-0046
Moriyama Y, Hamada H, Okada M, Tsuchiya N, Maru H, Shirato Y et al (2010) Distinctive clinical features of human bocavirus in children younger than 2 years. Eur J Pediatr 169(9):1087–1092. https://doi.org/10.1007/s00431-010-1183-x
Verbeke V, Reynders M, Floré K, Vandewal W, Debulpaep S, Sauer K et al (2019) Human bocavirus infection in Belgian children with respiratory tract disease. Arch Virol 164(12):2919–2930. https://doi.org/10.1007/s00705-019-04396-6
Abozahra R, Abdelhamid SM, Khairy K, Baraka K (2020) Detection and phylogenetic analysis of Human bocavirus in children diagnosed with acute respiratory tract infection. J Med Microbiol 69(9):1197–1202. https://doi.org/10.1099/jmm.0.001243
Petrarca L, Nenna R, Frassanito A, Pierangeli A, Di Mattia G, Scagnolari C et al (2020) Human bocavirus in children hospitalized for acute respiratory tract infection in Rome. World J Pediatr 16(3):293–298. https://doi.org/10.1007/s12519-019-00324-5
Pozo F, García-García ML, Calvo C, Cuesta I, Pérez-Breña P, Casas I (2007) High incidence of human bocavirus infection in children in Spain. J Clin Virol 40(3):224–228. https://doi.org/10.1016/j.jcv.2007.08.010
Redshaw WC, Rich F, Grimwood K, Kirman JR (2007) Human bocavirus in infants, New Zealand. Emerg Infect Dis 13:1797–1799. https://doi.org/10.3201/eid1311.070793
Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM (2006) Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol 78(9):1232–1240. https://doi.org/10.1002/jmv.20689
Nokso-Koivisto J, Pyles RB, Miller AL, Jennings K, Loeffelholz M, Chonmaitree T (2014) Role of human bocavirus in upper respiratory tract infections and acute otitis media. J Pediatr Infect Dis Soc 3(2):98–103. https://doi.org/10.1093/jpids/pit061
Regamey N, Frey U, Deffernez C, Latzin P, Kaiser L (2007) Isolation of human bocavirus from Swiss infants with respiratory infections. Pediatr Infect Dis J 26(2):177–179. https://doi.org/10.1097/01.inf.0000250623.43107.bc
Lambert SB, Ware RS, Cook AL, Maguire FA, Whiley DM, Bialasiewicz S et al (2012) Observational Research in Childhood Infectious Diseases (ORChID): a dynamic birth cohort study. BMJ Open 2(6):1–8. https://doi.org/10.1136/bmjopen-2012-002134
Sarna M, Ware RS, Lambert SB, Sloots TP, Nissen MD, Grimwood K (2018) Timing of first respiratory virus detections in infants: a community-based birth cohort study. J Infect Dis 217(3):418–427. https://doi.org/10.1093/infdis/jix599
Sarna M, Lambert SB, Sloots TP, Whiley DM, Alsaleh A, Mhango L et al (2018) Viruses causing lower respiratory symptoms in young children: findings from the ORChID birth cohort. Thorax 73(10):969–979. https://doi.org/10.1136/thoraxjnl-2017-210233
Alsaleh AN, Whiley DM, Bialasiewicz S, Lambert SB, Ware RS, Nissen MD et al (2014) Nasal swab samples and real-time polymerase chain reaction assays in community-based, longitudinal studies of respiratory viruses: the importance of sample integrity and quality control. BMC Infect Dis 14:15. https://doi.org/10.1186/1471-2334-14-15
O’Grady K-AF, Torzillo PJ, Rockett RJ, Whiley DM, Nissen MD, Sloots TP et al (2011) Successful application of a simple specimen transport method for the conduct of respiratory virus surveillance in remote Indigenous communities in Australia. Trop Med Int Health 16(6):766–772. https://doi.org/10.1111/j.1365-3156.2011.02757.x
Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM (2006) Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol 35(1):99–102. https://doi.org/10.1016/j.jcv.2005.09.008
Zoch-Lesniak B, Ware RS, Grimwood K, Lambert SB (2020) The respiratory specimen collection trial (ReSpeCT): a randomized controlled trial to compare quality and timeliness of respiratory sample collection in the home by parents and healthcare workers from children aged <2 years. J Pediatr Infect Dis Soc 9(2):134–141. https://doi.org/10.1093/jpids/piy136
Christensen A, Dollner H, Skanke LH, Krokstad S, Moe N, Nordbo SA (2013) Detection of spliced mRNA from human bocavirus 1 in clinical samples from children with respiratory tract infections. Emerg Infect Dis 19(4):574–580. https://doi.org/10.3201/eid1904.121775
Meriluoto M, Hedman L, Tanner L, Simell V, Mäkinen M, Simell S et al (2012) Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg Infect Dis 18(2):264–271. https://doi.org/10.3201/eid1802.111293
Powell E, Sumner E, Shaw AG, Calvez R, Fink CG, Kroll JS (2022) The temporal pattern and lifestyle associations of respiratory virus infection in a cohort study spanning the first two years of life. BMC Pediatr 22(1):166. https://doi.org/10.1186/s12887-022-03215-3
Azziz-Baumgartner E, Bruno A, Daugherty M, Chico ME, Lopez A, Arriola CS et al (2022) Incidence and seasonality of respiratory viruses among medically attended children with acute respiratory infections in an Ecuador birth cohort, 2011–2014. Influenza Other Respir Viruses 16(1):24–33. https://doi.org/10.1111/irv.12887
Victora CG, Bahl R, Barros AJD, França GVA, Horton S, Krasevec J et al (2016) Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet 387(10017):475–490. https://doi.org/10.1016/S0140-6736(15)01024-7
Kusel MMH, de Klerk NH, Holt PG, Kebadz T, Johnston SL, Sly PD (2006) Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life. A birth cohort study. Pediatr Infect Dis J 25(8):680–686. https://doi.org/10.1097/01.inf.0000226912.88900.a3
von Linstow M-L, Holst KK, Larsen K, Koch A, Andersen PK, Høgh B (2008) Acute respiratory symptoms and general illness during the first year of life: a population-based birth cohort study. Pediatr Pulmonol 43(6):584–593. https://doi.org/10.1002/ppul.20828
de Jong BM, van der Ent CK, van Putte KN, van der Zalm MM, Verheij TJM, Kimpen JLL et al (2007) Determinants of health care utilization for respiratory symptoms in the first year of life. Med Care 45(8):746–752. https://doi.org/10.1097/MLR.0b013e3180546879
de Hoog MLA, Venekamp RP, van der Ent CK, Schilder A, Sanders EA, Damoiseaux RA et al (2014) Impact of early daycare on healthcare resource use related to upper respiratory tract infections during childhood: prospective WHISTLER cohort study. BMC Med 12(1):107. https://doi.org/10.1186/1741-7015-12-107
Moriyama M, Hugentobler WJ, Iwasaki A (2020) Seasonality of respiratory viral infections. Annu Rev Virol 7(1):83–101. https://doi.org/10.1146/annurev-virology-012420-022445
Morley C, Grimwood K, Maloney S, Ware RS (2018) Meteorological factors and respiratory syncytial virus seasonality in subtropical Australia. Epidemiol Infect 146(6):757–762. https://doi.org/10.1017/S0950268818000614
Takashima MD, Grimwood K, Sly PD, Lambert SB, Ware RS (2022) Interference between rhinovirus and other RNA respiratory viruses in the first 2-years of life: a longitudinal community-based birth cohort study. J Clin Virol 155:105249. https://doi.org/10.1016/j.jcv.2022.105249
Saha S, Grimwood K, Lambert SB, Ware RS (2020) Parainfluenza virus infection in an Australian community-based birth cohort. Pediatr Infect Dis J 39(9):e284–e287. https://doi.org/10.1097/INF.0000000000002796
Grimwood K, Lambert SB, Ware RS (2020) Endemic non-SARS-CoV-2 human coronaviruses in a community-based Australian birth cohort. Pediatrics 146(5).e2020009316. https://doi.org/10.1542/peds.2020-009316
Takashima MD, Grimwood K, Sly PD, Lambert SB, Chappell KJ, Watterson D et al (2021) Epidemiology of respiratory syncytial virus in a community birth cohort of infants in the first 2 years of life. Eur J Pediatr 180(7):2125–2135. https://doi.org/10.1007/s00431-021-03998-0
Emerson J, Cochrane E, McNamara S, Kuypers J, Gibson RL, Campbell AP (2013) Home self-collection of nasal swabs for diagnosis of acute respiratory virus infections in children with cystic fibrosis. J Pediatr Infect Dis Soc 2(4):345–351. https://doi.org/10.1093/jpids/pit039
Woodall CA, Thornton H V., Anderson EC, Ingle SM, Muir P, Vipond B et al (2021) Prospective study of the performance of parent-collected nasal and swab samples, compared with nurse-collected swab samples, for the molecular detection of respiratory microorganisms. Microbiol Spectr 9(3). https://doi.org/10.1128/Spectrum.00164-21
Author information
Authors and Affiliations
Contributions
SBL, RSW, and KG contributed to the study concept and design. SS, NF, SBL, RSW, and KG contributed to data extraction and/or collection. SS, NF, and RSW undertook the data analysis and had access to all data. All authors contributed to the data interpretation. SS drafted the manuscript with supervision and revisions contributed to by SBL, RSW, and KG. All authors approved the final version of the paper.
Corresponding author
Ethics declarations
Ethics approval
The Children’s Health Queensland (HREC/10/QRCH/16), the Royal Brisbane and Women’s Hospital (HREC/10/QRBW125), and The University of Queensland (2010000820) Human Research Ethics Committees approved the study.
Consent to participate
Written informed consent was obtained from the parents.
Competing interests
The ORChID study was supported by an Australian National Health and Medical Research Council (NHMRC) project grant (GNT615700) and a programme grant (5006) from the Children’s Hospital Foundation (CHF). SBL was the recipient of an NHMRC Early Career Fellowship and a CHF Mid-career Fellowship. SBL, RSW, and KG have received NHMRC grants for this and other projects. SS and NF have no conflict of interest to declare.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saha, S., Fozzard, N., Lambert, S.B. et al. Human bocavirus-1 infections in Australian children aged < 2 years: a birth cohort study. Eur J Clin Microbiol Infect Dis 42, 99–108 (2023). https://doi.org/10.1007/s10096-022-04529-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-022-04529-x