Abstract
Context
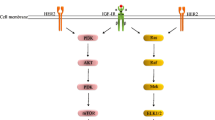
Epidermal growth factor receptor (EGFR), a member of the HER receptor family, is over expressed in various cancer cells. Using tumor-specific antibodies to deliver cytotoxic agents directly to the tumor cells is an effective treatment strategy. Targeted therapy by fusing anti-EGFR scFv with tumor-specific cytokines promises the emergence of a new era.
Methods
We designed a novel immuno-apoptotic fusion protein, anti-EGFR scFv-IL-24, consisting of a specific cancer cell targeting antibody and recombinant cytokine IL-24 to explore its anti-cancerous potential. Amino acid sequences of both anti-EGFR scFv and IL-24 were fused using a specific rigid linker. In silico characterization of the designed fusion protein like to predict the primary, secondary, physiochemical properties, quality, and structural validation using online bioinformatic tools. The newly designed fusion protein consists of 402 amino acids that showed good quality with a predicted value of 76.7% having 81.5% residues in the most favored region as predicted by ERRAT2 and Ramachandran plot analysis. Docking and simulation studies were performed using HDOCK and Desmond module of Schrodinger. All the parameters of quality, validity, interaction analysis, and stability suggested that the fused molecule is fully operational and functional. The results of the study support that the anti-EGFR scFv-IL-24 fused protein could be proved as a novel candidate to combat cancer.








Similar content being viewed by others
References
Jones PA, Baylin SBJC (2007) The epigenomics of cancer 128(4):683–692
Frank SA (2018) Dynamics of cancer. Princeton University Press
Hassanpour SH, Dehghani M (2017) Review of cancer from perspective of molecular. J Cancer Res Pract 4(4):127–129
Schrack JA, Gresham G, Wanigatunga AA (2017) Understanding physical activity in cancer patients and survivors: new methodology, new challenges, and new opportunities. Mol Case Stud 3(4):a001933
Chen H et al (2017) Rethinking Cancer Nanotheranostics 2(7):1–18
Vazhappilly CG et al (2021) Current methodologies to refine bioavailability, delivery, and therapeutic efficacy of plant flavonoids in cancer treatment 94:108623
Hegde PS, Chen DS (2020) Top 10 challenges in cancer immunotherapy. Immunity 52(1):17–35
Waldmann TA (2018) Cytokines in cancer immunotherapy. Cold Spring Harbor Perspect Biol. 10(12):a028472
Smith LL et al (2000) Chemoprevention of breast cancer by tamoxifen: risks and opportunities. 30(5):571-594
Morris R, Kershaw NJ, Babon JJ (2018) The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci 27(12):1984–2009
Menezes ME et al (2014) MDA-7/IL-24: multifunctional cancer killing cytokine. In: Grimm S (ed) Anticancer Genes. Springer, London, London, pp 127–153
Nielsen TO et al (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. 10(16):5367-5374
Brandes AA et al (2008) Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. 14(4):957-960
Hatanpaa KJ et al (2010) Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. 12(9):675–684
Zhou Y et al (2012) Impact of intrinsic affinity on functional binding and biological activity of EGFR antibodiescancer cell targeting of anti-EGFR antibodies. 11(7):1467-1476
Weisser NE, Hall JC (2009) Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotecnol Adv 27(4):502–520
Nguyen PV et al (2020) Targeted nanomedicine with anti-EGFR scFv for siRNA delivery into triple negative breast cancer cells. 157:74-84
Ma C et al (2012) Synthesis and purification of a toxin-linked conjugate targeting epidermal growth factor receptor in Escherichia coli. 83(1):1-7
Nautiyal K et al (2019) Design and assessment of an active anti-epidermal growth factor receptor (EGFR) single chain variable fragment (ScFv) with improved solubility. 508(4):1043–1049
David JM et al (2017) A novel bifunctional anti-PD-L1/TGF-β Trap fusion protein (M7824) efficiently reverts mesenchymalization of human lung cancer cells. 6(10):e1349589
Hutmacher C, Neri D (2019) Antibody-cytokine fusion proteins: biopharmaceuticals with immunomodulatory properties for cancer therapy. Adv Drug Deliv Rev 141:67–91
Le Joncour V, Laakkonen P (2018) Seek & destroy, use of targeting peptides for cancer detection and drug delivery. Bioorganic Med Chem 26(10):2797–2806
Chen X et al (2012) Pharmacokinetics of recombinant bifunctional fusion proteins. 8(5):581-595
Reddy Chichili VP, Kumar V (2013) Sivaraman J (2013) Linkers in the structural biology of protein–protein interactions. Protein Sci 22(2):153–167
Sen TZ et al (2005) GOR V server for protein secondary structure prediction. 21(11):2787-2788
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405
Rezaie E et al (2020) Bioinformatics predictions, expression, purification and structural analysis of the PE38KDEL-scfv immunotoxin against EPHA2 receptor. 26:979–996
Zhang J et al (2009) Design and optimization of a linker for fusion protein construction. Prog Nat Sci 19(10):1197–1200
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:1–8
Yang J et al (2015) The I-TASSER suite: protein structure and function prediction. 12(1):7-8
Bowie JU, Lüthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253(5016):164–170
Ko J et al (2012) GalaxyWEB server for protein structure prediction and refinement. 40(W1):W294-W297
Gasteiger E et al (2005) Protein identification and analysis tools on the ExPASy server. Springer
Hebditch M et al (2017) Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics 33(19):3098–3100
Sharma N, et al (202) AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief Bioinform 22(4)
Sharma N, et al (2022) ToxinPred2: an improved method for predicting toxicity of proteins. Brief Bioinform 23(5)
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8(1):4
Yan Y et al (2017) HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 45(W1):W365–W373
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40(1):82–92
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graphics 14(1):33–38
Phillips JC, et al (2020) Scalable molecular dynamics on CPU and GPU architectures with NAMD. 153(4):044130
Case DA, et al (2021) Amber 2021: Reference Manual
Case DA, et al (2005) The Amber biomolecular simulation programs. 26(16):1668-1688
Jorgensen W, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926
Duan Y, et al (2003) A point‐charge force field for molecular mechanics simulations of proteins based on condensed‐phase quantum mechanical calculations. 24(16):1999-2012
Grant BJ, Skjærven L, Yao XQ (2021) The Bio3D packages for structural bioinformatics. Protein Sci 30(1):20–30
Hebditch M, et al (2017) Protein–Sol: a web tool for predicting protein solubility from sequence. 33(19):3098–3100
Yan Y, et al (2017) HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Rese 45
Funding
This work was supported in part by grants from the HEC Pakistan under NRPU 2021 (Project No. 16935) and the University of the Punjab Lahore Pakistan.
Author information
Authors and Affiliations
Contributions
Methodology, formal analysis, validation: Z, H.B, N.Y; investigation resources: S.A; data curation: S.Q, Z, N.Y; and original draft preparation: Z, H.B, N.Y, S.A, S.Q. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zaroon, yousaf, N., Aslam, S. et al. In silico investigation of a novel anti-EGFR scFv-IL-24 fusion protein induces apoptosis in malignant cells. J Mol Model 29, 282 (2023). https://doi.org/10.1007/s00894-023-05690-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05690-6