Abstract
Context
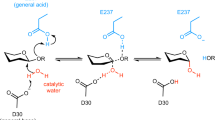
The putative endoglucanase, PsGH5A, from Pseudopedobacter saltans of family GH5 contains a catalytic module, PsGH5 (β/α)8-TIM barrel), at N-terminal followed by a family 6 carbohydrate-binding module (CBM6, β-sandwich). Superposition of PsGH5A with PDB homologs revealed Glu220 and Glu318 as evolutionarily conserved and catalytic residues performing the hydrolysis through retaining-type mechanism, a canonical property of GH5 family. PsGH5A showed higher affinity for longer cellooligosaccharides, as long as cellodecaose with binding free energy (∆G) of − 13.72 kcal/mol upon the molecular docking, thereby indicating the endo-mode of hydrolysis. The radius of gyration, Rg (2.7 nm), and solvent accessible surface area, SASA (229.6 nm2), of PsGH5A-Cellotetraose complex were determined by MD simulation which was lower than that of PsGH5A (Rg, 2.8 nm, SASA, 267 nm2) demonstrating the compactness and affinity of PsGH5A with the cellulosic ligands. Cellulose compatibility of PsGH5A was further confirmed by MMPBSA and per-residue decomposition analysis, where notable ∆G of − 54.38 kcal/mol for PsGH5A-Cellotetraose complex was observed. Thus, PsGH5A could be potentially an efficient endoglucanase as it accommodated larger cellooligosaccharides at its active-site. PsGH5A is the first putative endoglucanase studied here from P. saltans which could be genome-mined for lignocellulosic biomass saccharification in the renewable energy sector.
Methods
The 3-D structure of PsGH5A generated by AlphaFold2, RaptorX, SwissModel, Phyre2 and Robetta tool; YASARA was used for energy minimization of built models. UCLA SAVES-v6 was used for quality assessment of models. Molecular Docking was performed using SWISS-DOCK server and Chimera software. Molecular Dynamics simulations and MMPBSA analysis of PsGH5A and PsGH5A-Cellotetraose complex were performed on GROMACS 2019.6.
Graphical abstract











Similar content being viewed by others
Data availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EC:
-
Enzyme commission
- CBH:
-
Cellobiohydrolase
- PSI-BLAST:
-
Position-Specific Iterative Basic Local Alignment Search Tool
- TIM:
-
Triose phosphate isomerase
- AA:
-
Amino acids
- NCBI:
-
National Center for Biotechnology Information
- EMBL:
-
European Molecular Biology Laboratory
- PDB:
-
Protein Data Bank
- UCLA:
-
University of California
- HTC:
-
High-throughput computing
- NVT:
-
Normal Volume Temperature
- NPT:
-
Normal Pressure Temperature
- RMSD:
-
Root Mean Square Deviation
- RMSF:
-
Root Mean Square Fluctuation
- SD:
-
Standard Dviation
- MMPBSA:
-
Molecular Mechanics Poisson-Boltzmann Surface Area
References
Singh S, Dhillon A, Goyal A (2020) Enhanced catalytic efficiency of Bacillus amyloliquefaciens SS35 endoglucanase by ultraviolet directed evolution and mutation analysis. Renew Energy 151:1124–1133. https://doi.org/10.1016/j.renene.2019.11.105
Gavande PV, Nath P, Kumar K et al (2022) Highly efficient, processive and multifunctional recombinant endoglucanase RfGH5_4 from Ruminococcus flavefaciens FD-1 v3 for recycling lignocellulosic plant biomasses. Int J Biol Macromol 209:801–813
Cantarel BL, Coutinho PM, Rancurel C et al (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:233–238. https://doi.org/10.1093/nar/gkn663
Drula E, Garron M-L, Dogan S et al (2022) The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50:D571–D577
Gavande PV, Kumar K, Ahmed J, Goyal A (2022) Multifunctionality and mechanism of processivity of family GH5 endoglucanase, RfGH5_4 from Ruminococcus flavefaciens on lignocellulosic polymers. Int J Biol Macromol 224:1395–1411
Thakur A, Sharma K, Jamaldheen SB, Goyal A (2020) Molecular characterization, regioselective and synergistic action of first recombinant type III α-L-arabinofuranosidase of family 43 glycoside hydrolase (PsGH43_12) from Pseudopedobacter saltans. Mol Biotechnol 62:443–455
Nath P, Goyal A (2021) Structure and dynamics analysis of multi-domain putative β-1,4-glucosidase of family 3 glycoside hydrolase (PsGH3) from Pseudopedobacter saltans. J Mol Model 27:1–16
Sharma K, Antunes IL, Rajulapati V, Goyal A (2018) Low-resolution SAXS and comparative modeling based structure analysis of endo-β-1, 4-xylanase a family 10 glycoside hydrolase from Pseudopedobacter saltans comb. nov. Int J Biol Macromol 112:1104–1114
Sharma K, Morla S, Khaire KC et al (2020) Extraction, characterization of xylan from Azadirachta indica (neem) sawdust and production of antiproliferative xylooligosaccharides. Int J Biol Macromol 163:1897–1907
Thakur A, Sharma A, Khaire KC et al (2021) Two-step saccharification of the xylan portion of sugarcane waste by recombinant xylanolytic enzymes for enhanced xylose production. ACS Omega 6:11772–11782
Liolios K, Sikorski J, Lu M et al (2011) Complete genome sequence of the gliding, heparinolytic Pedobacter saltans type strain (113T). Stand Genomic Sci 5:30–40
Benson DA, Clark K, Karsch-Mizrachi I et al (2015) GenBank. Nucleic Acids Res 43:D30
Sussman JL, Lin D, Jiang J et al (1998) Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr Sect D Biol Crystallogr 54:1078–1084
Boratyn GM, Camacho C, Cooper PS et al (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33
Lu S, Wang J, Chitsaz F et al (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268
Ren J, Wen L, Gao X et al (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19:271–273
Laskowski RA, Jabłońska J, Pravda L et al (2018) PDBsum: structural summaries of PDB entries. Protein Sci 27:129–134
Wu X, Wan X-F, Wu G et al (2006) Phylogenetic analysis using complete signature information of whole genomes and clustered neighbour-joining method. Int J Bioinform Res Appl 2:219–248
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11:681–684
Klose DP, Janes RW (2010) 2Struc-the Protein Secondary Structure Analysis Server. Biophys J 98:454a–455a
Källberg M, Margaryan G, Wang S et al (2014) RaptorX server: a resource for template-based protein structure modeling. In: Protein structure prediction. Springer, vol 1137, pp 17–27
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Park H, Kim DE, Ovchinnikov S et al (2018) Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins Struct Funct Bioinforma 86:283–291
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589
Land H, Humble MS (2018) YASARA: a tool to obtain structural guidance in biocatalytic investigations. In: Protein engineering. Springer, vol 1685, pp 43–67
Kumar K, Singh S, Sharma K, Goyal A (2021) Computational modeling and small-angle X-ray scattering based structure analysis and identifying ligand cleavage mechanism by processive endocellulase of family 9 glycoside hydrolase (HtGH9) from Hungateiclostridium thermocellum ATCC 27405. J Mol Graph Model 103:107808
Laskowski RA, MacArthur MW, Thornton JM (2006) PROCHECK: validation of protein-structure coordinates. International Tables for Crystallography In: Crystallography of biological macromolecules Wiley, vol. F. ch. 25.2, pp 722–725
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
Singh S, Ahmed J, Gavande PV et al (2023) Structural and functional insights into the glycoside hydrolase family 30 xylanase of the rumen bacterium Ruminococcus flavefaciens. J Mol Struct 1272:134155
Hess B, Kutzner C, Van Der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Grosdidier A, Zoete V, Michielin O (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39:W270–W277
Kuttel MM, Ståhle J, Widmalm G (2016) CarbBuilder: software for building molecular models of complex oligo‐and polysaccharide structures. J Comput Chem 37(22):2098–2105
Pettersen EF, Goddard TD, Huang CC et al (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82
Visualizer BDS (2021) Dassault Systemes; San Diego, CA, USA: 2021. Version v21 1:20298
Yuan S, Chan HCS, Hu Z (2017) Using PyMOL as a platform for computational drug design. Wiley Interdiscip Rev Comput Mol Sci 7:e1298
Medeiros DDJ, Cortopassi WA, Costa França TC, Pimentel AS (2013) ITP adjuster 1.0: a new utility program to adjust charges in the topology files generated by the PRODRG server. J Chem 2013(803151):1–6
Jain M, Muthukumaran J, Singh AK (2021) Comparative structural and functional analysis of STL and SLL, chitin-binding lectins from Solanum spp. J Biomol Struct Dyn 39:4907–4922
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Kumari R, Kumar R, Consortium OSDD, Lynn A (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962
Cao J, Lai Q, Li G, Shao Z (2014) Pseudopedobacter beijingensis gen. nov., sp. nov., isolated from coking wastewater activated sludge, and reclassification of Pedobacter saltans as Pseudopedobacter saltans comb. nov. Int J Syst Evol Microbiol 64:1853–1858
Garg VK, Avashthi H, Tiwari A et al (2016) MFPPI–multi FASTA ProtParam interface. Bioinformation 12:74
Dos Santos CR, Cordeiro RL, Wong DWS, Murakami MT (2015) Structural basis for xyloglucan specificity and α-d-Xyl p (1→ 6)-d-Glcp recognition at the −1 subsite within the GH5 family. Biochemistry 54:1930–1942
Hollingsworth SA, Karplus PA (2010) A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. BioMol Concepts 1(3–4):271–283
Dym O, Eisenberg D, Yeates TO (2012) ERRAT. International Tables for Crystallography In: Crystallography of biological macromolecules Wiley, Vol. F. ch. 21.3, pp. 678–679
Lombard V, Golaconda Ramulu H, Drula E et al (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495
Domı́nguez R, Souchon H, Lascombe M-B, Alzari PM (1996) The crystal structure of a family 5 endoglucanase mutant in complexed and uncomplexed forms reveals an induced fit activation mechanism. J Mol Biol 257:1042–1051
Cutfield JF, Sullivan PA, Cutfield SM (2000) Minor structural consequences of alternative CUG codon usage (Ser for Leu) in Candida albicans exoglucanase. Protein Eng 13:735–738
Zheng F, Ding S (2013) Processivity and enzymatic mode of a glycoside hydrolase family 5 endoglucanase from Volvariella volvacea. Appl Environ Microbiol 79:989–996
Kumari R, Dhankhar P, Dalal V (2021) Structure-based mimicking of hydroxylated biphenyl congeners (OHPCBs) for human transthyretin, an important enzyme of thyroid hormone system. J Mol Graph Model 105:107870. https://doi.org/10.1016/J.JMGM.2021.107870
Acknowledgements
PVG is thankful to the Ministry of Education (erstwhile, Ministry of Human Resource Development (MHRD), Govt. of India, for the fellowship through Graduate Aptitude Test for Engineering (GATE). The authors also thank Param-Ishan Supercomputing facility at IIT Guwahati for molecular simulations.
Funding
The research work was supported by a Trilateral project with the Indian Institute of Technology Bombay, Mumbai, India and All India Institute of Medical Sciences (AIIMS), New Delhi, India under the NERBPMC DBT-Twinning project scheme with the grant (No. BT/PR24786/NER/95/853/2017) from Department of Biotechnology, Ministry of Science and Technology, New Delhi, Government of India to AG.
Author information
Authors and Affiliations
Contributions
Parmeshwar Vitthal Gavande: methodology, investigation, visualization, software and data curation, formal analysis, validation, writing—original draft preparation, rewriting and editing. Arun Goyal: conceptualization, supervision, resources, project administration, reviewing, rewriting, editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This research did not involve any human participants or animals to generate and analyse the data used in this study.
Conflict of interest
The authors declare no competing interests for regarding this work.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• PsGH5A contains canonical (β/α)8-TIM barrel for catalytic module.
• Associated CBM6 module possesses β-sandwich fold.
• PsGH5A with Glu220 and Glu318 catalytic residues shows retaining-type mechanism.
• MD simulation of PsGH5A-Cellotetraose complex showed stability and compactness.
• PsGH5A active-site can accommodate cellulosic ligands as large as cellodecaose.
• ∆G by MMPBSA indicated higher affinity of PsGH5A for cellulosic ligands.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gavande, P.V., Goyal, A. Molecular dynamics–based structural insights of the first putative endoglucanase, PsGH5A of glycoside hydrolase family 5 from Pseudopedobacter saltans. J Mol Model 29, 186 (2023). https://doi.org/10.1007/s00894-023-05575-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05575-8